A large number of projects in the RKM lab are focused on dissecting the mechanisms of epigenetic regulation using Next Generation Sequencing (NGS) technologies. The team at BIC employs computational and bioinformatics-based approaches to perform the large-scale data analysis that these projects entail. NGS project management including technical and experimental design of the projects, quality control checks, and data management and tracking are provided for a seamless end-to-end service. We also aim to provide bench biologists better access to computational solutions by developing user friendly and interactive tools and databases for data visualization and analysis.
Key Research Interests
Chromatin State and Epigenomics
Accessibility of chromatin in different cell types plays a crucial role in controlling the functional outcome of the DNA
sequence via epigenetic marks. These include posttranslational modifications of histones, DNA methylation
and association with chromatin modulator proteins. We employ ChIP-seq, RNA-seq and bisulphite sequencing
techniques to investigate genome-wide changes during physiological processes such as aging, stress response,
transgenerational inheritance and development and differentiation. We also perform meta-analyses using existing
datasets of various genomes from public repositories.
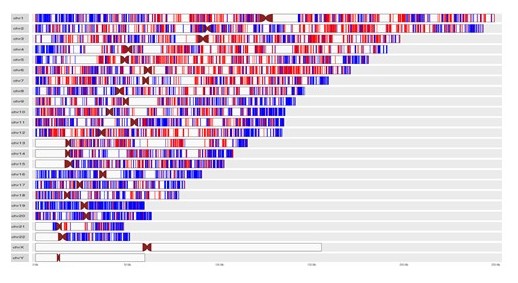
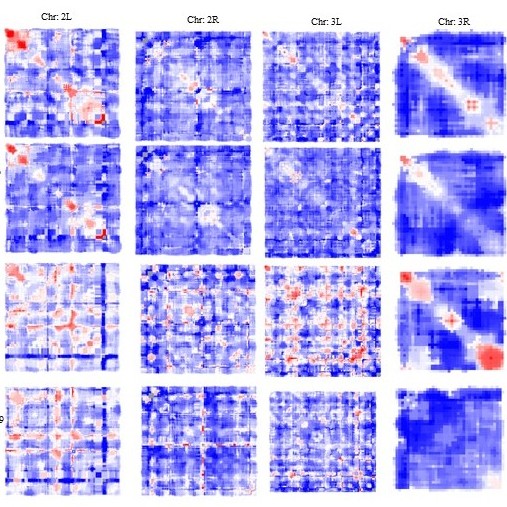
Nuclear Architecture and Genome Organization
Chromatin in the eukaryotic nucleus is organized in the form of loops and domains which are anchored to the Nuclear Matrix
(NuMat) via DNA elements known as Matrix Associated Regions (MARs). The NuMat additionally contains RNA and
proteins to provide a structural and functional backbone for the nuclear processes. We investigate the identity
and developmental dynamics of NuMat components via MAR and NuMat RNA sequencing to understand the functional
organization of the nucleus during mitosis, development and disease.

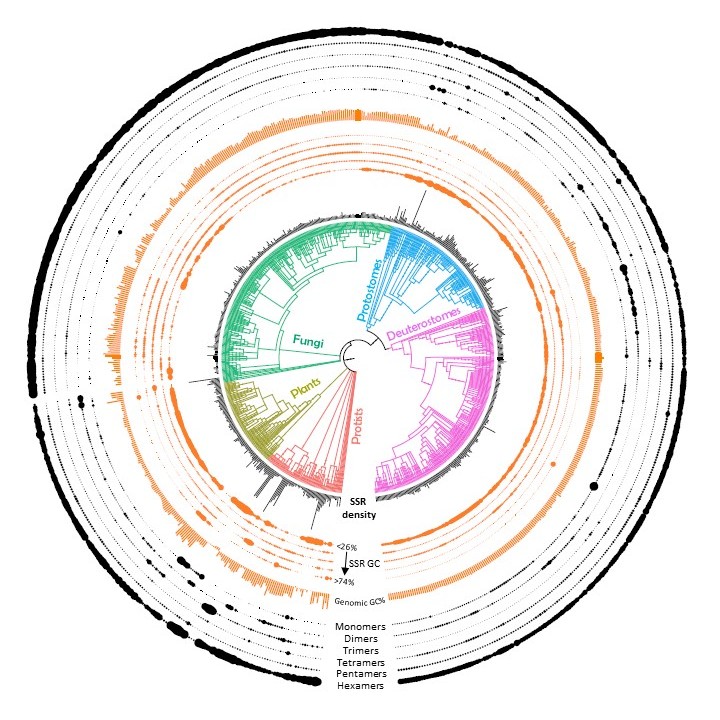
Simple Sequence Repeats
Microsatellites, or Simple Sequence Repeats (SSRs), are conserved repeat elements distributed non-randomly in all genomes.
Contrary to earlier beliefs of their propagation in genomes as “junk DNA”, they have been shown to perform
functional roles in gene regulation and we believe that they are critical elements in evolution and speciation.
We have recently generated an exhaustive collection of >650 million SSRs from ~7000 genomes that can be explored
using our
interactive database (MSDB). We are currently involved in investigating their role in genome organization
as well as dissecting their evolutionary trends to reconstruct phylogenetic relationships.
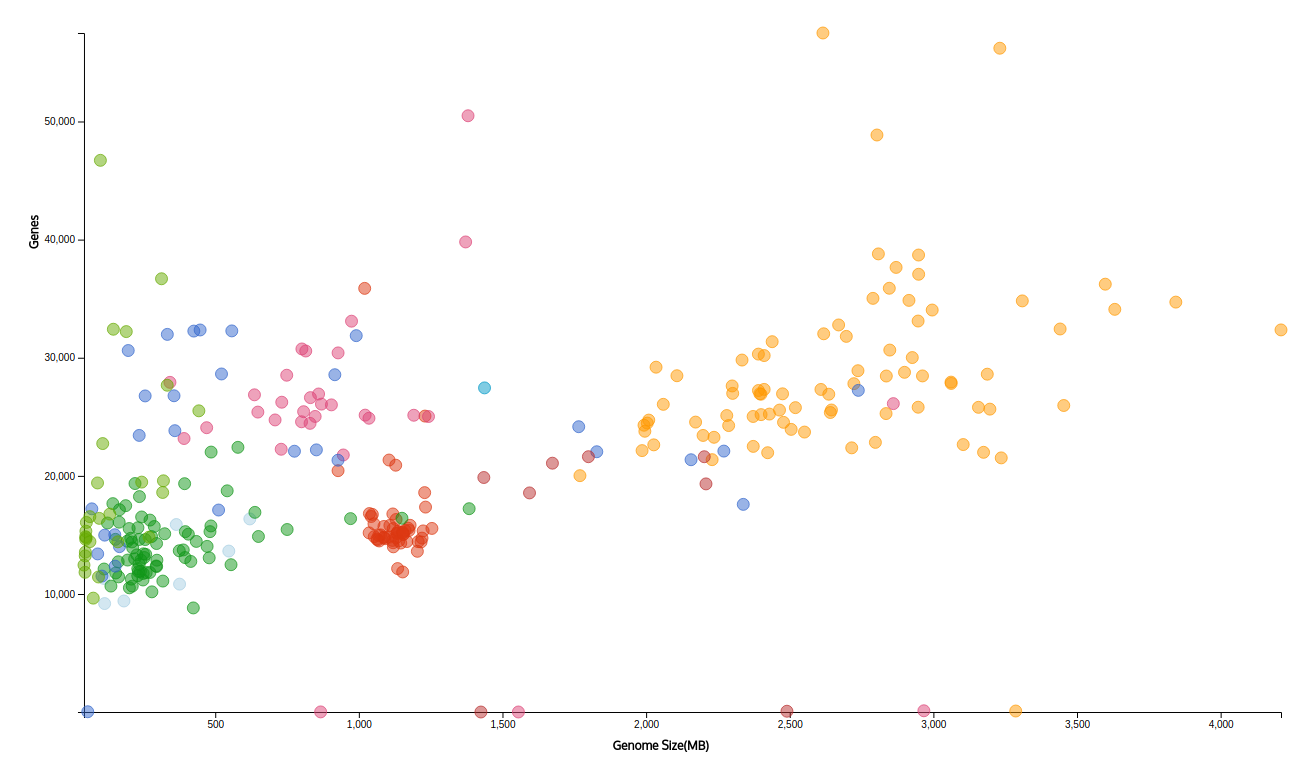
De novo genome sequencing
Genome information is crucial to understanding conserved gene regulatory mechanisms and aspects of body design, especially
regulation of the homeotic genes for development of the anterior-posterior axis. We are invested in generating
the genome and transcriptome resources of species that are of interest in our lab. We have recently sequenced
and assembled the genome of the parasitic wasp,
Leptopilina boulardi, using a combination of Illumina and PacBio sequencing.
