SnS-Align (Structure and Sequence Alignment) is a tool developed in perl, used to align the protein sequences by combining sequence and structure information at an instance. Given a protein sequence of an organism, the tool is used to identify its homologue(s) in the evolutionarily distant organisms, which might not be found from either sequence alone or structure alone comparisons. A snapshot of the tool is illustrated below.
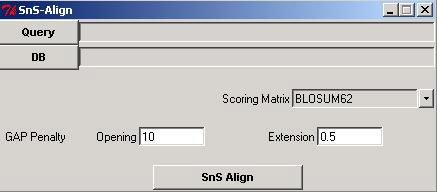
Query should be a FASTA formatted protein sequence file. DB (Database) is a set of mutliple protein sequences in FASTA format. You can see the SnS-algnment in a Graphical User interface. *(star) indicates the conservation of secondary structure and + (plus) indicates the conservation of sequence. You can save the alignment in Text format or Post script format by selecting the save alignment option.
Requirements:
-Perl is to be installed with the Tk module (use Active-Perl for windows and for linux get your rpm)
-EMBOSS is to be installed to run the program (EMBOSS for Windows and Linux is freely available)
Download
SnS-Align is available for Windows and Linux
For further queries contact mishra@ccmb.res.in |