| 
|
|
| Elife (2018) DOI: 10.7554/eLife.38232 |
|
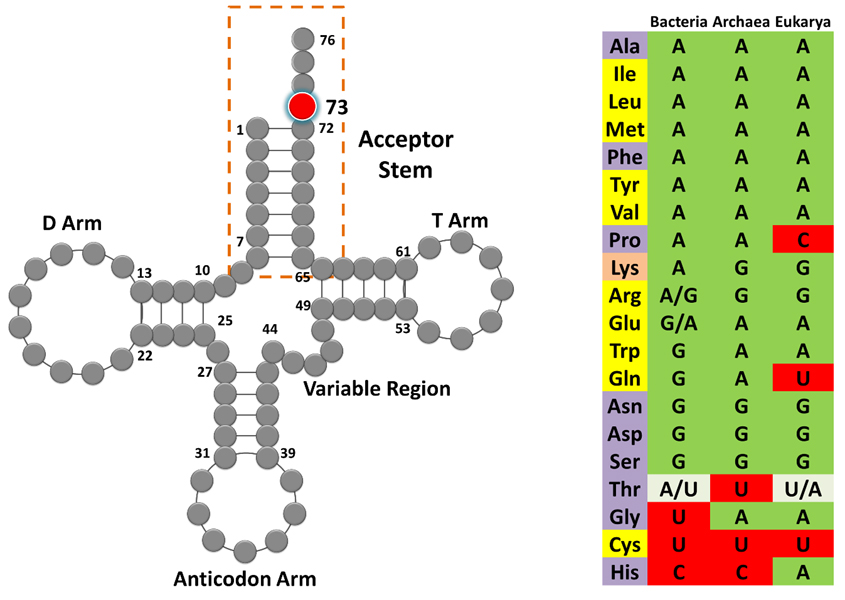
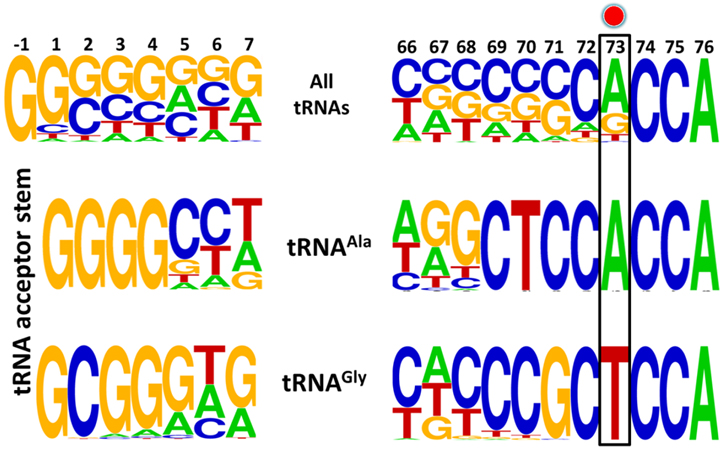
tRNAGly and tRNAAla show discriminator base dichotomy in Bacteria. (a) Clover leaf model of tRNA with the discriminator base highlighted in red.
Distribution of the discriminator base in all tRNAs across the three domains of life.
|
Frequency distribution of tRNA acceptor stem elements across bacterial tRNAs, comparing and contrasting between tRNAGly and tRNAAla. Red circle indicates the discriminator base. |
|
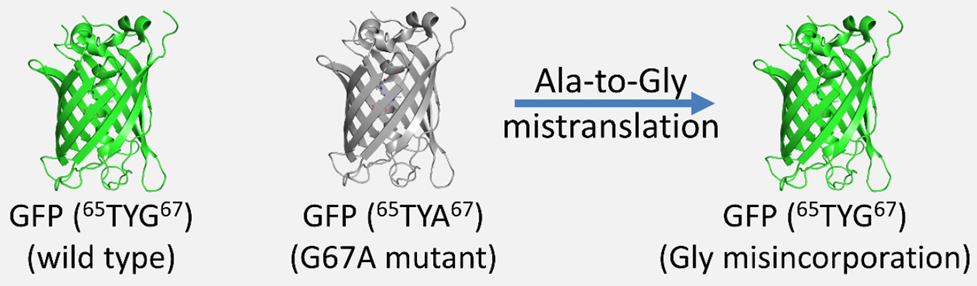
DTD avoids glycine misincorporation into proteins. (a) GFP-based fluorescence reporter assay for
visualizing alanine-to-glycine mistranslation, wherein the mutant GFP G67A (65TYA67) will fluoresce only when TYA is mistranslated to TYG.
|
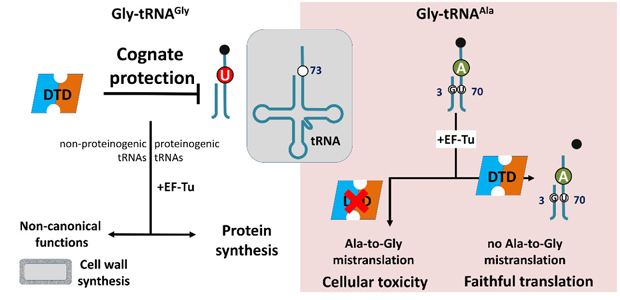
Model showing N73 dichotomy in bacterial tRNAGly and tRNAAla, enabling protection of the cognate Gly-tRNAGly (both proteinogenic and non-proteinogenic) predominantly by U73, while effecting efficient removal of the non-cognate Gly-tRNAAla (having A73 and G3.U70) to prevent alanine-to-glycine mistranslation. |
|
Nature Communications (2018) 9 : 511, doi:10.1038/s41467-017-02204-w
PDB –ID: 5XAQ |
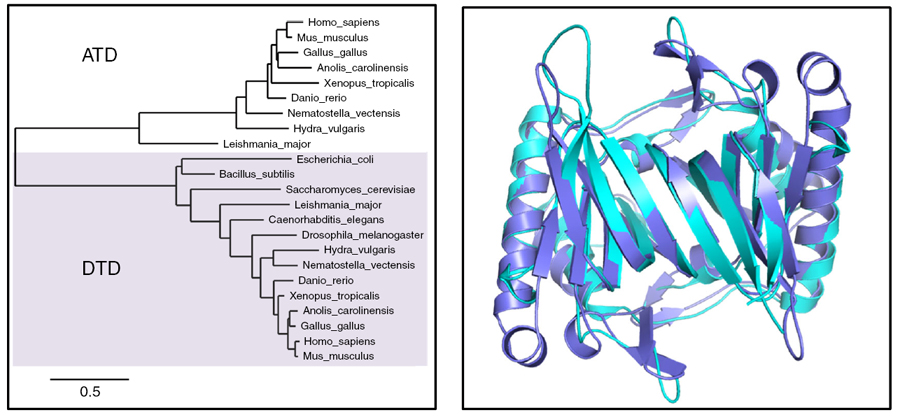
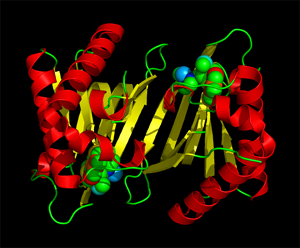
ATD belongs to the DTD-like fold
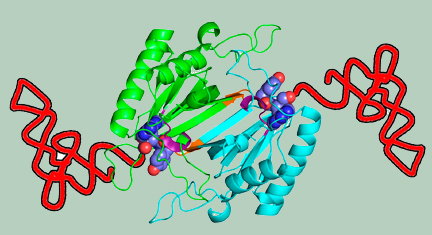
Crystal structure of MmATD homodimer (blue) superimposed on that of PfDTD homodimer (cyan; PDB id: 4NBI) |
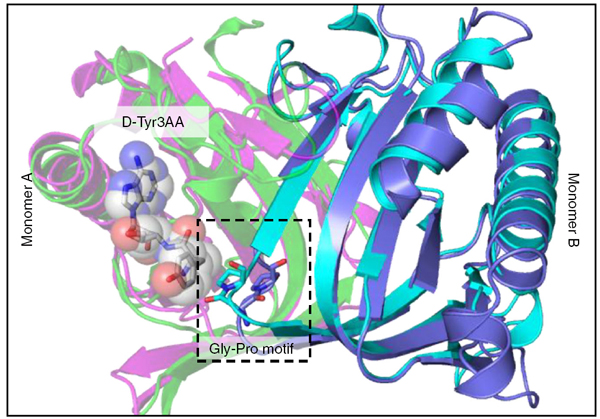
Structural superposition of MmATD on PfDTD displaying the cross-subunit Gly-Pro motif |
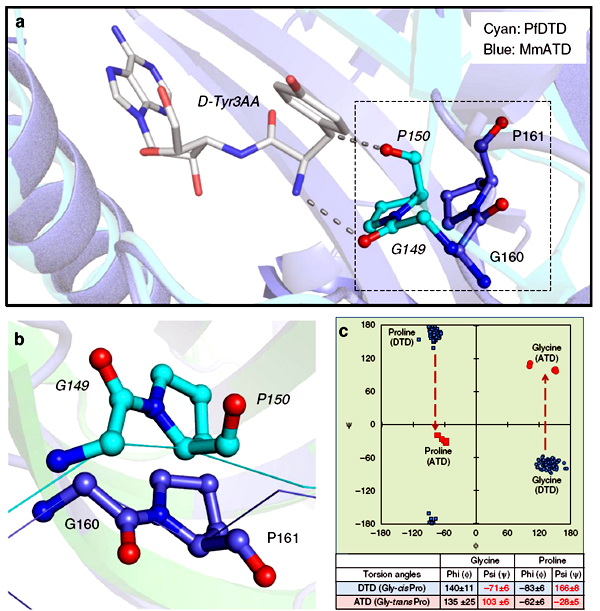
ATD has a Gly-transPro motif in the active site, unlike a Gly-cisPro motif in DTD |
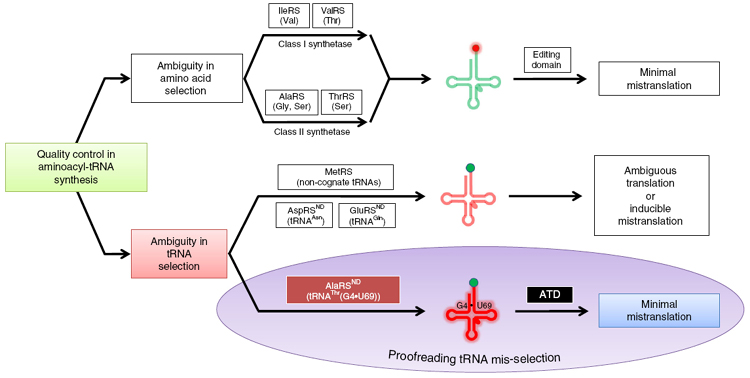
ATD is a unique and dedicated proofreading factor that rectifies a critical tRNA selection error. Model for mis-selection and consequent misacylation of tRNAThr(G4•U69) with L-alanine by AlaRSND, and its subsequent proofreading by ATD. |
|
| |
| Elife(2017) |
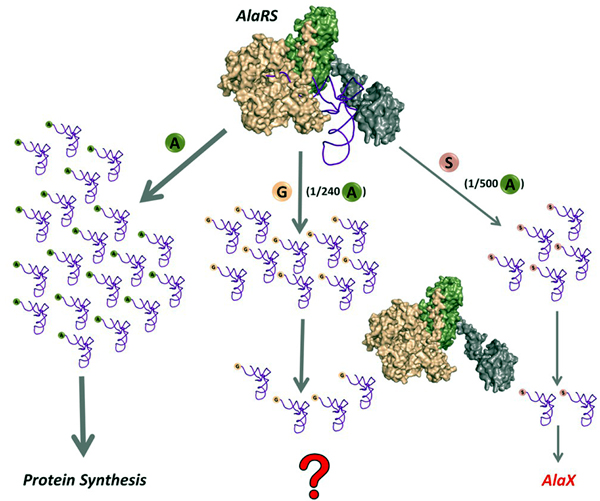
Mischarging Glycine and Serine by AlaRS |
Misacylation of tRNAAla with glycine by AlaRS and its prevention/rectification by DTD. |
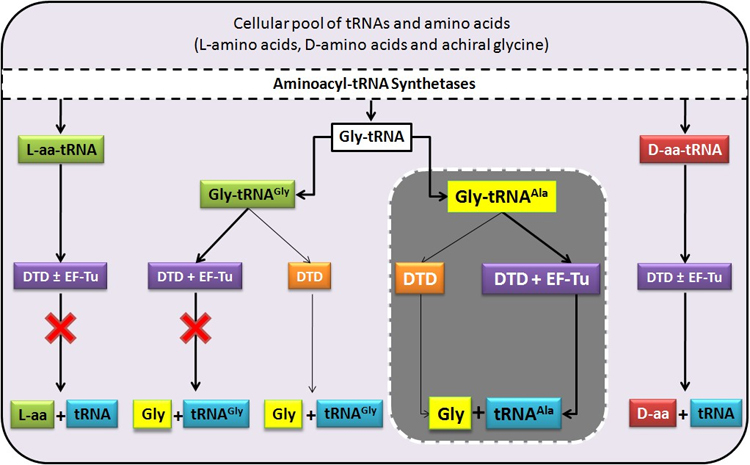
DTD doubles as a key factor to uncouple glycine mischarged on tRNAAla. |
|
| |
Misediting of Achiral Glycine by D-Aminoacyl-tRNA Deacylase
PLOS Biology | DOI:10.1371/journal.pbio.1002465
PDB-ID: 5J61 |
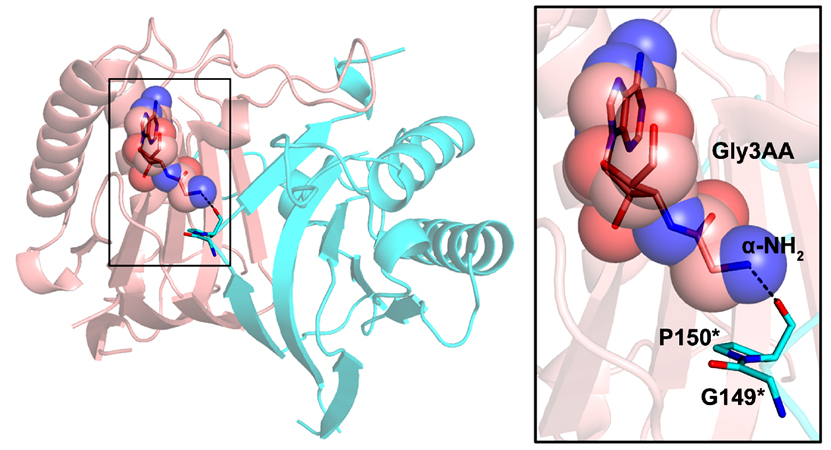
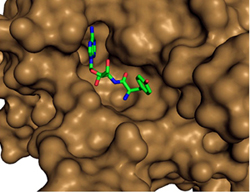
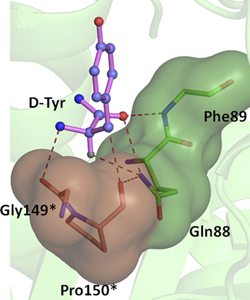
Crystal structure of PfDTD with Gly3AA |
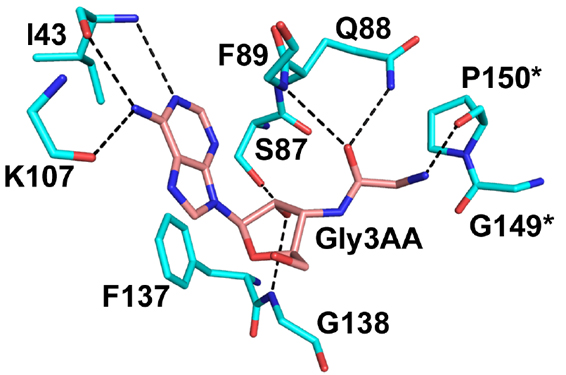
Network of interactions of Gly3AA with active site residues of PfDTD. *Residues from the dimeric counterpart |
|
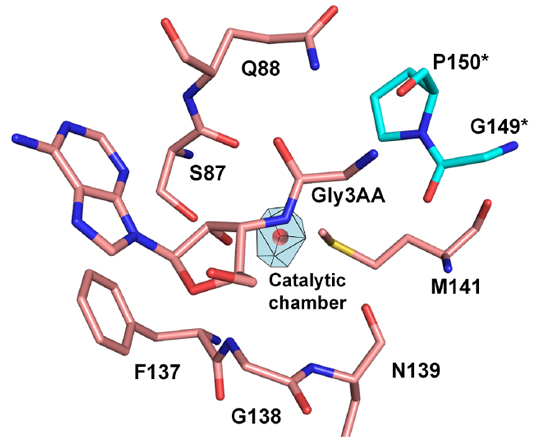
Catalytic chamber in the active site of PfDTD can accommodate a water molecule |
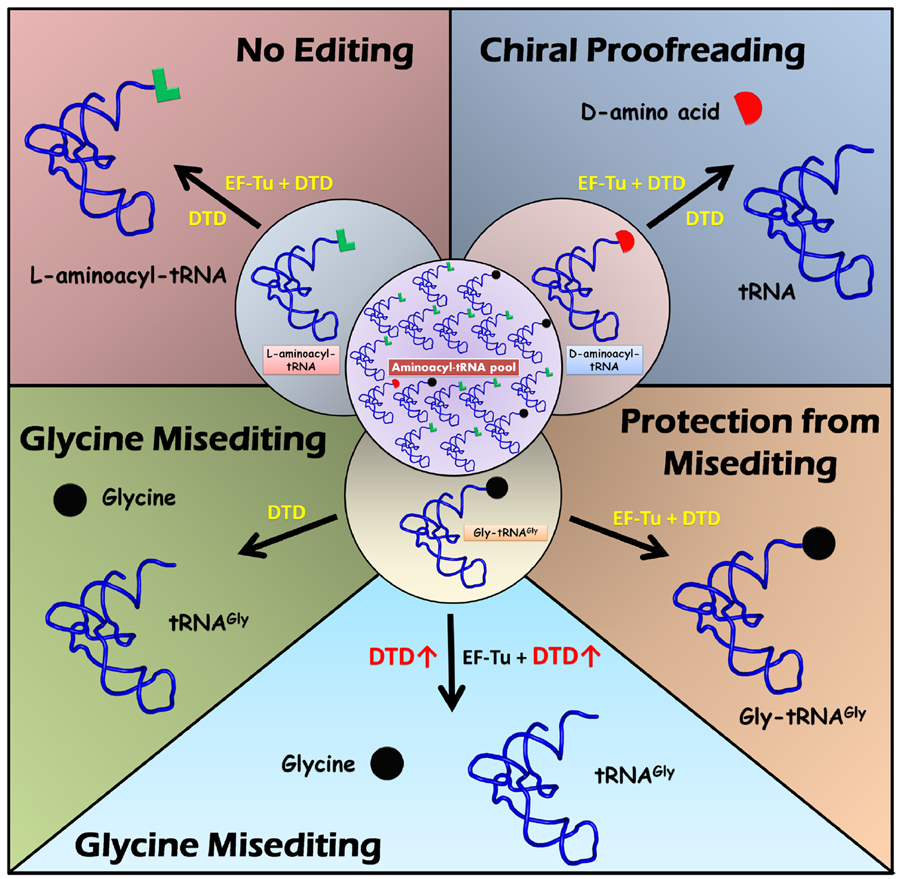
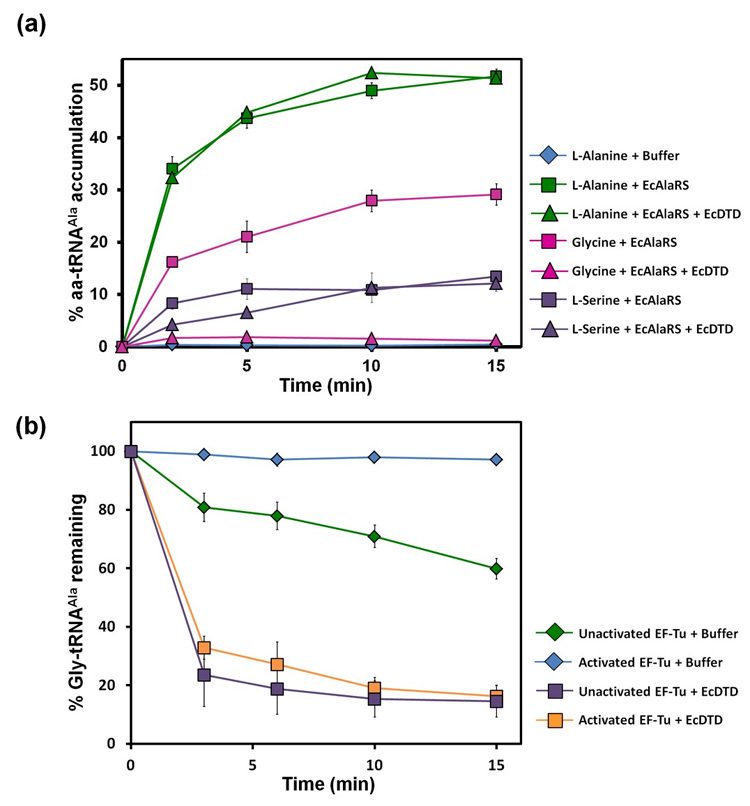
Model for protection of Gly-tRNAGly by EF-Tu |
|
| |
Nature Communications (2015) 6 : 7552, doi:10.1038/ncomms8552
PDB-IDs: 4RR6, 4RR7, 4RR8, 4RR9, 4RRA, 4RRB, 4RRC, 4RRD, 4RRF, 4RRG, 4RRH, 4RRI, 4RRJ, 4RRK, 4RRL, 4RRM, 4RRQ and 4RRR |
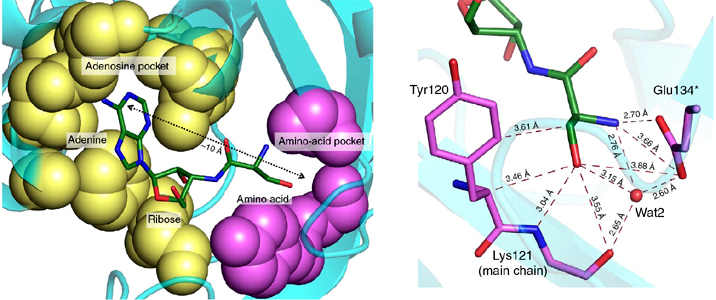
Specificity and catalysis hardwired at the RNA–protein interface in a translational proofreading enzyme |
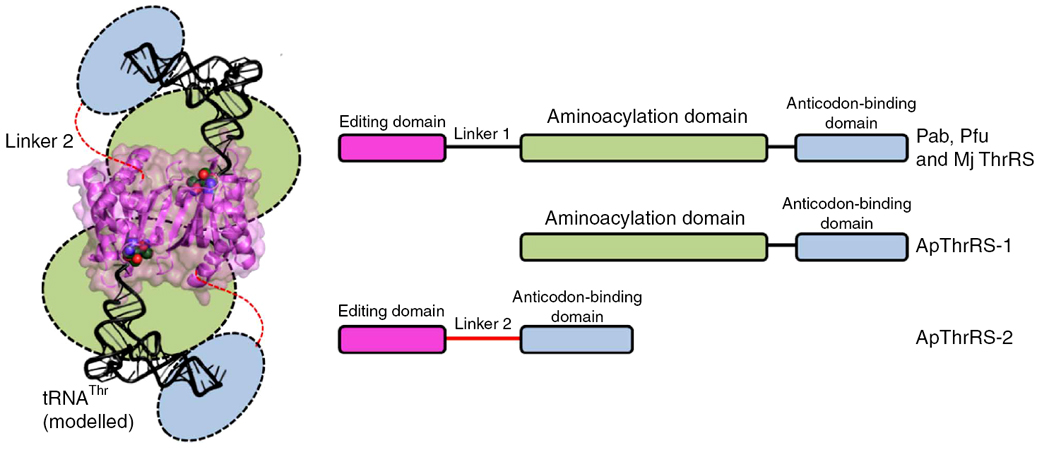
Domain organization of archaeal threonyl-tRNA synthetase (ThrRS) |
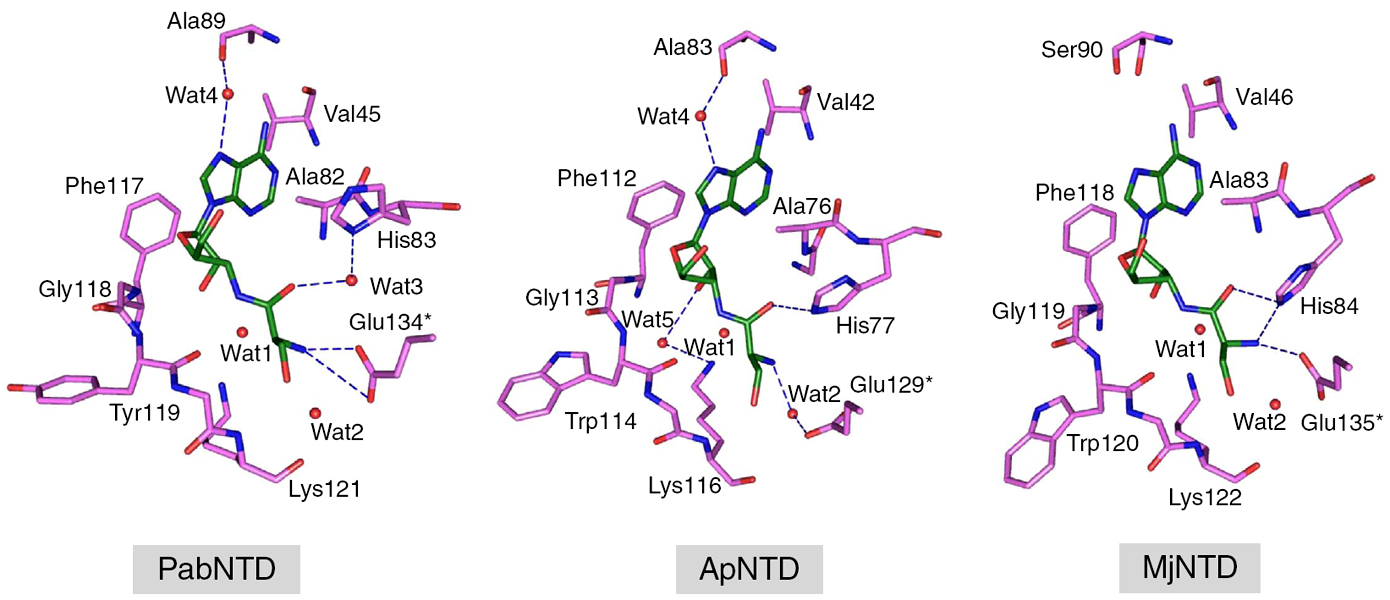
Plasticity in substrate recognition modes in NTD |
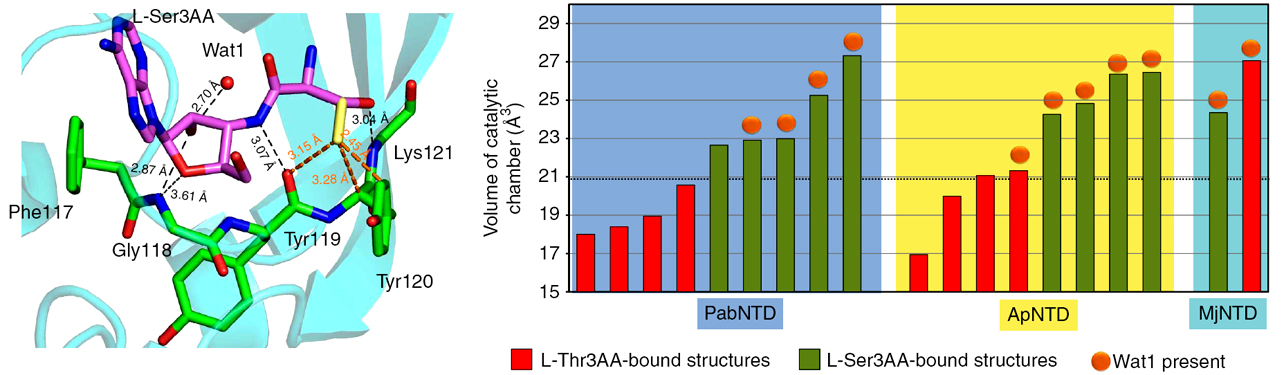
Role of the RNA–protein interface in substrate specificity |
|
| |
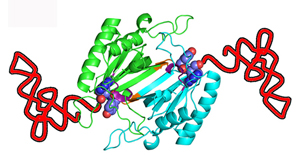
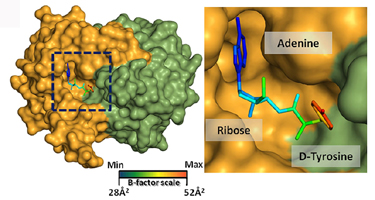
D-aminoacyl-tRNA deacylase in complex with D-tyrosyl-3'-aminoadenosine
in Plasmodium falciparum
Elife (2013) 2: e01519-e01519
PDB-IDs : 4NBI, 4NBJ |
|
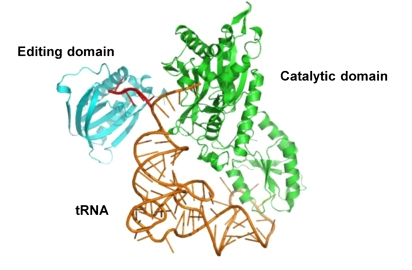
Archaeal ThrRS with tRNA
Thr |
2Fo–Fc map of post-transfer analog
|
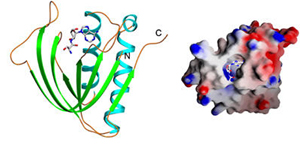
Pab-NTD–Ser3AA |
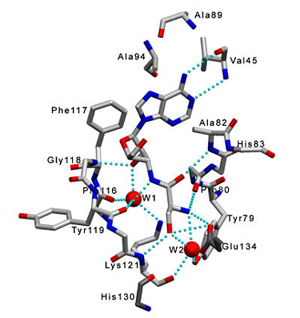
Ser3AA in the
active site |
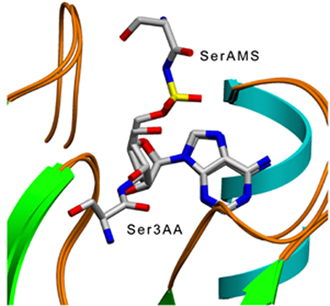
Structural
superposition of SerAMS and Ser3AA complex structures |
2HKZ
Pab-NTD–L-serine
|
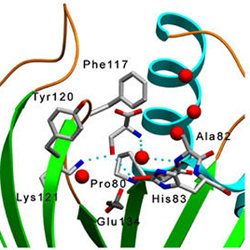
Residues interacting with L-serine at the active site pocket |
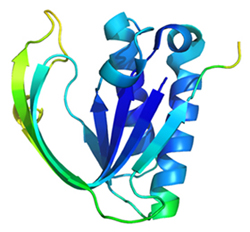
1Y2Q
Pab-NTD: Editing domain of threonyl-tRNA synthetase
|
|