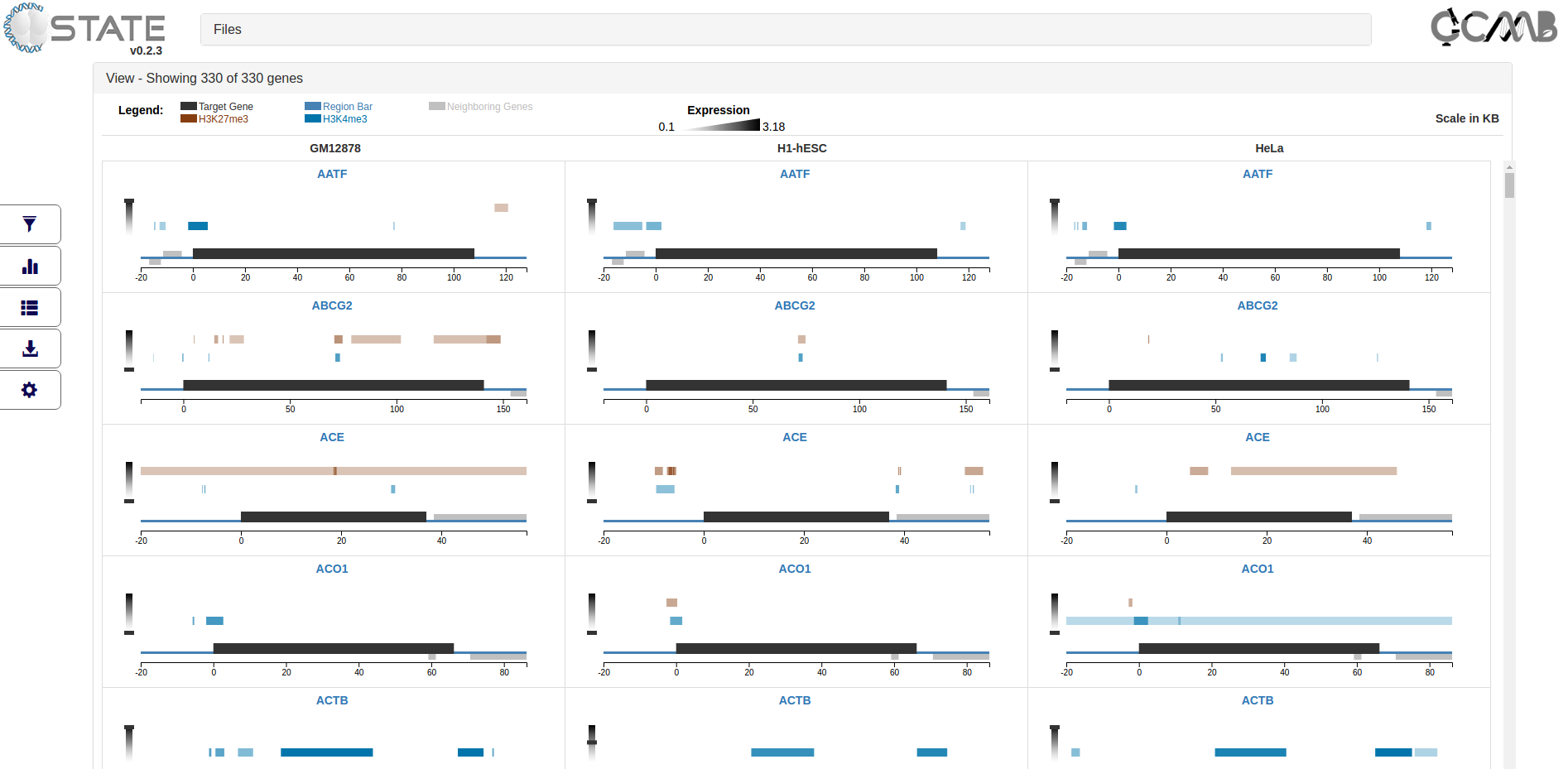
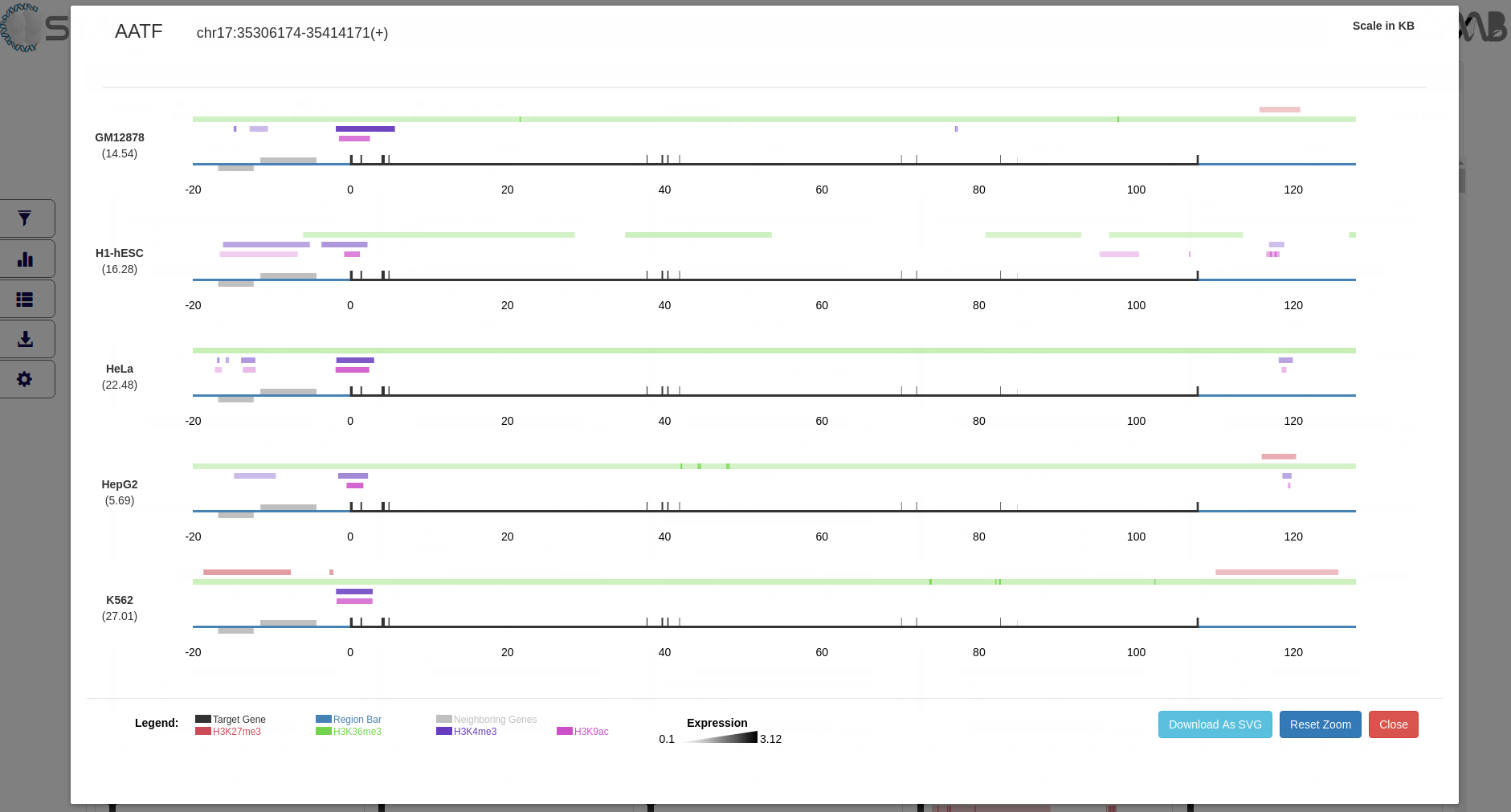
Data Analysis and Output
5 buttons on the left side of the Files and View accordions provide access to other functionalities of C-State.
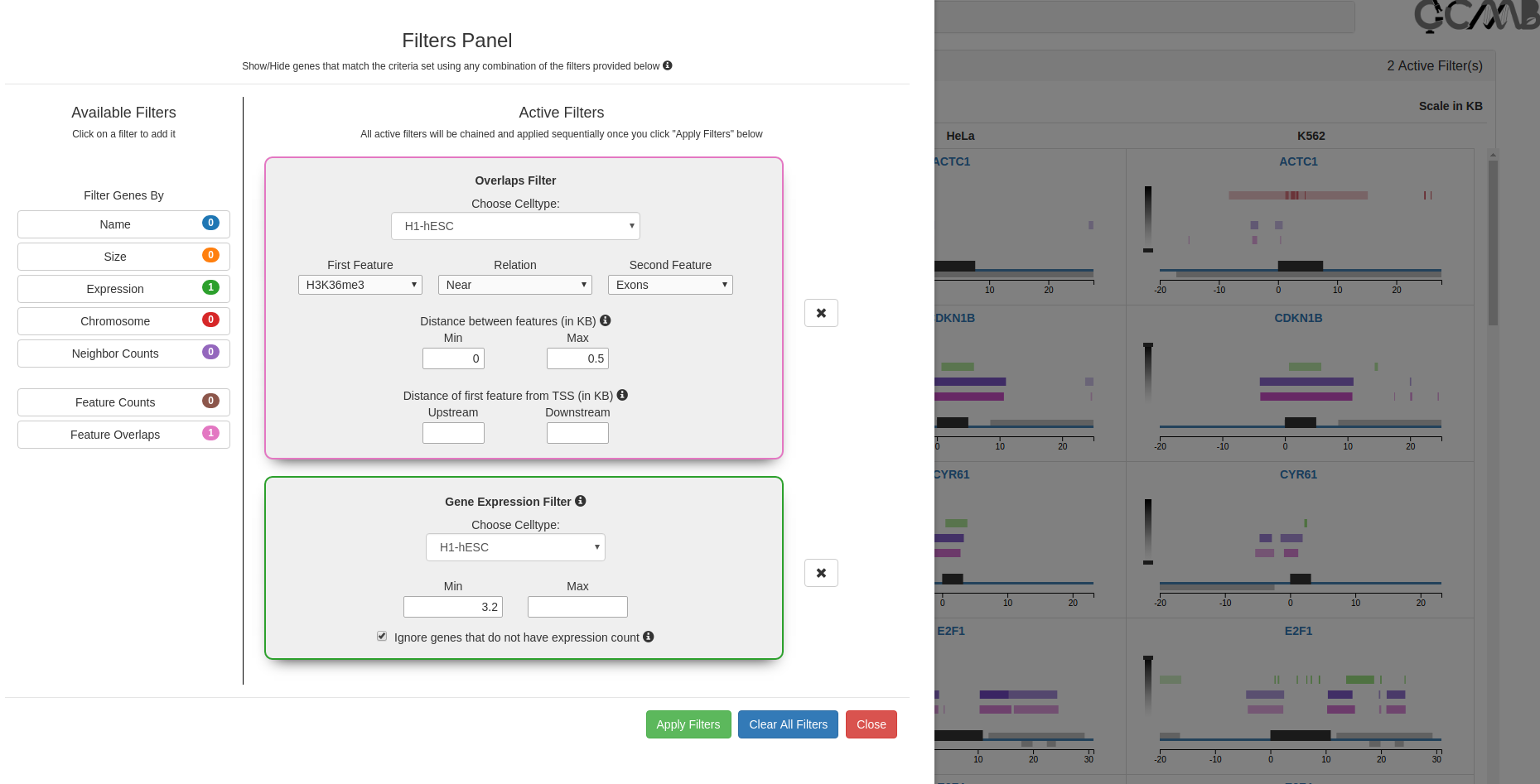
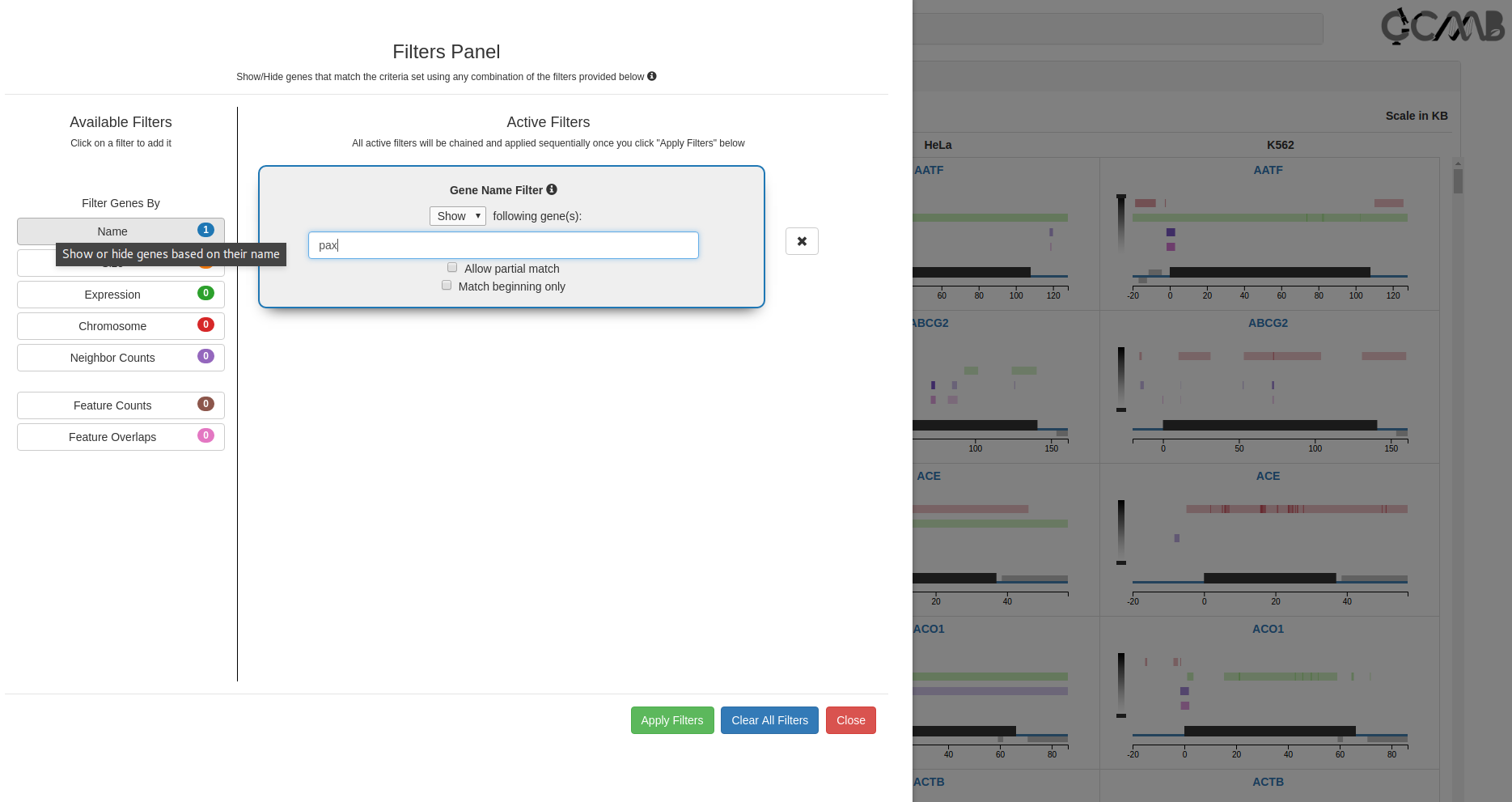
Filters Panel - Search for Epigenetic Patterns
One of the novel advantages of C-State is that it enables filtering and searching for cell type specific epigenetic
features and patterns. This is particularly useful for biologists to identify gene or cell type specific epigenetic
patterns without any need for programming or bioinformatics.
The filters module (1st button in the control panel) allows the user to search for genes containing specified patterns of
genomic and/or epigenetic features. Options in the filters appear automatically in the drop down menus based on the
raw data files provided to C-State. The filters can be chained together in the order of their selection to build
complex user-defined pattern searches since each output gets applied to the successive filters in the chain on clicking
the “Apply Filters” button (see use case examples).
Gene Name Filter
Search for a gene by name or ID; a set of gene names can be also be pasted to display all the genes that match
the given name(s). Ticking the 'Allow partial match’ or 'Match beginning only' checkboxes allows the user to
filter for all the genes which have similar names (eg., gene families such as Hox, PAX etc).
Gene Size Filter
Specify size cutoffs for genes or the total regions (including flanks) using a range of operators provided in
the drop down.
Gene Expression Filter
Search for genes/transcripts that fall within a given expression range in a cell type.
Chromosome Filter
Show or hide genes belonging to a particular chromosome. Chaining multiple chromosome filters allows viewing genes across
any chromosome combinations or across all chromosomes. This filter can also be used to display precise co-ordinates
of a locus.
Neighbor Counts Filter
Search for genes according to the presence of neighboring genes around them using a range of operators. User can optionally
choose to ignore or filter the dataset based on presence of other genes overlapping the target genes.
Apart from the above genomic filters that search based on gene size, location or genomic context, C-State also
provides options to identify genes based on their epigenetic patterns.
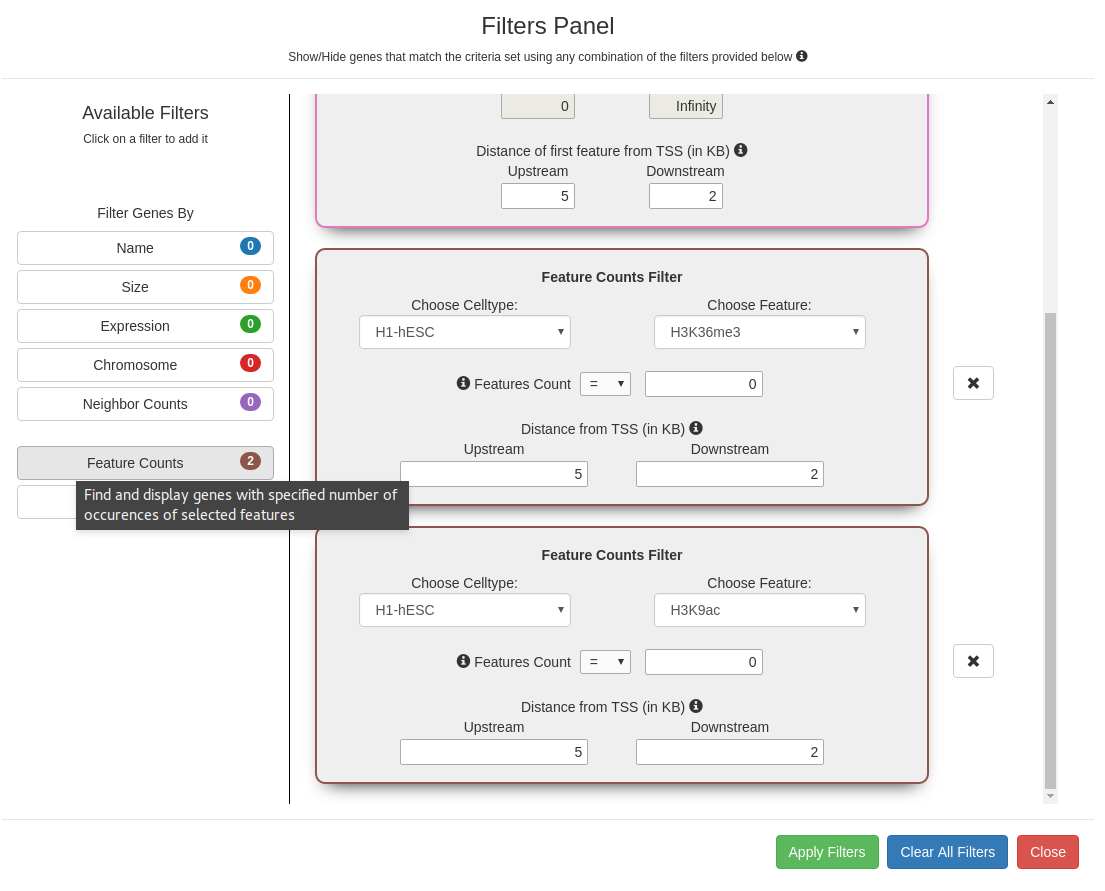
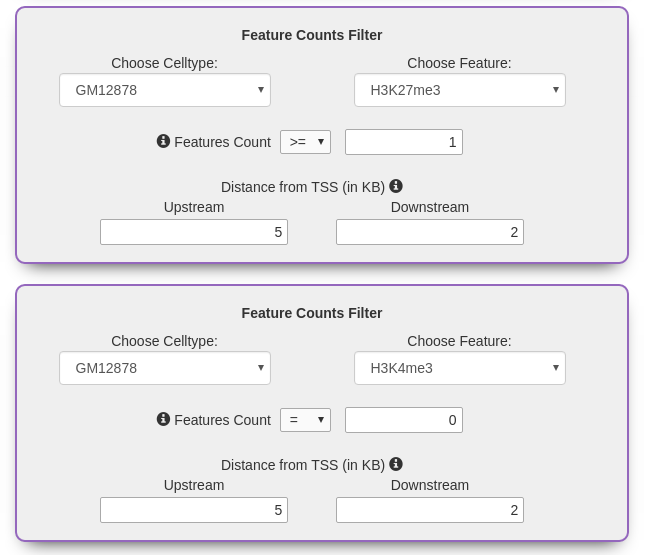
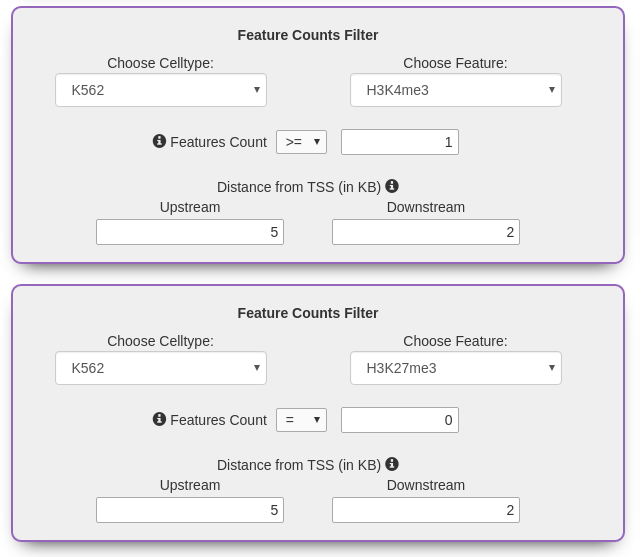
Feature Counts Filter
Select from a set of operators to find genes that carry the desired number of cell type specific marks. The search
can be further refined by specifying the distance of the marks from the TSS.
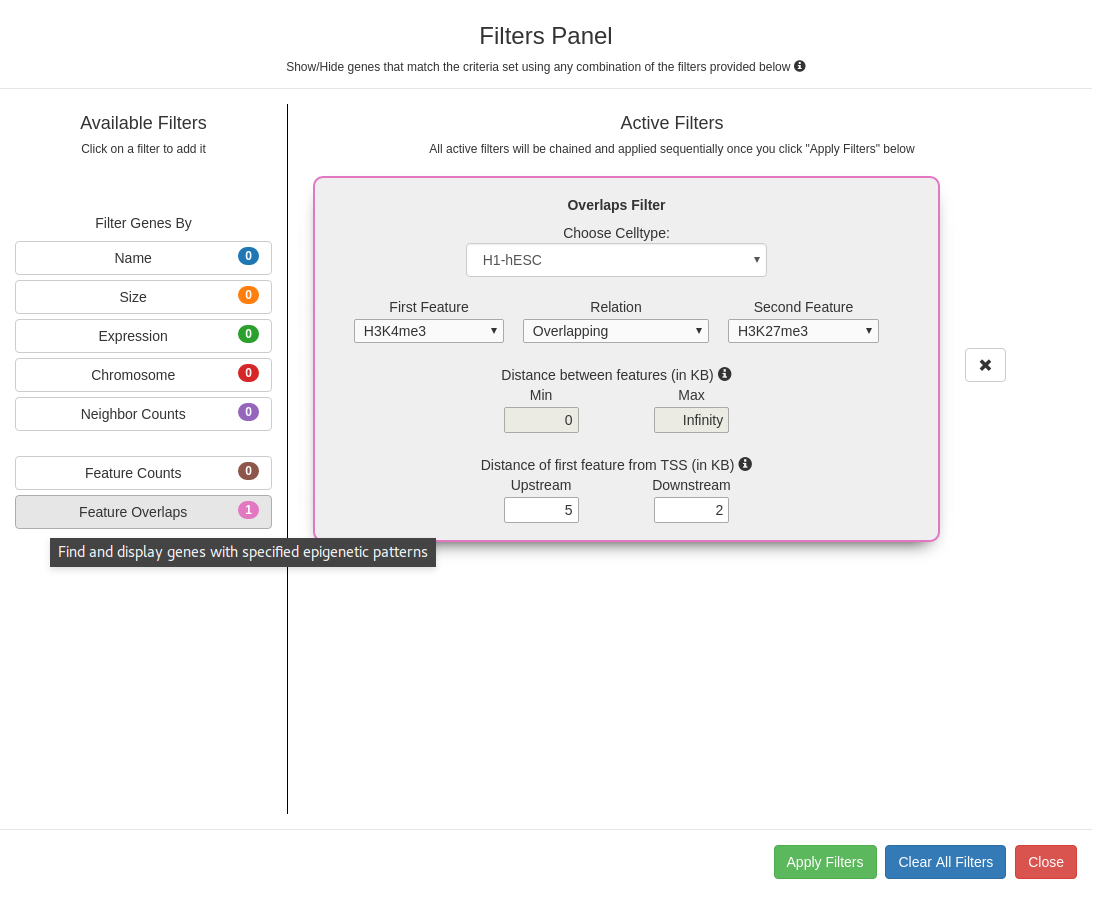
Feature Overlaps Filter
Filter to display genes based on cell type specific patterns of epigenetic marks. Select the two features that are to be
viewed in the context of each other from the two drop-down menus and define their relationship using the relation
dropdown:
- Upstream To - The first feature is present upstream to the second feature
- Downstream To - The first feature is present downstream to the second feature
- Near - The first feature can be either upstream or downstream to the second feature
- Overlapping - The first feature overlaps the second feature
The distance allowed between the two marks can also be specified; in case of overlaps, the min and max distance allowed between
the two marks is 0 and this option gets disabled. The search can be further refined by specifying the distance
of the pattern from the TSS.
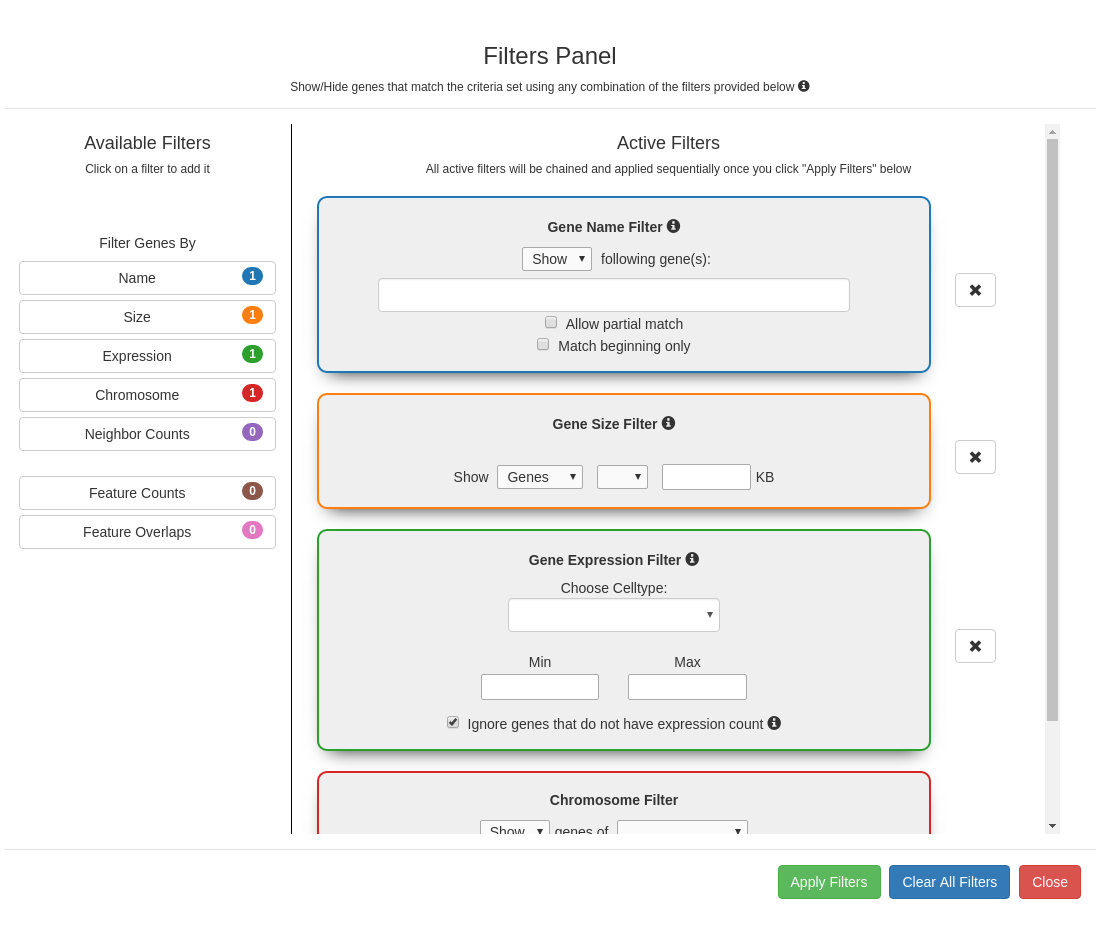 Filters panel showing the 7 available filters (Gene expression, genomic, and pattern filters). Clicking on a filter opens it
in the Active Filters area for setting parameters. Successive filters get chained sequentially; clicking on the
x next to a filter removes it from the chain. The filter combination is applied only on clicking the "Apply filters"
button.
Filters panel showing the 7 available filters (Gene expression, genomic, and pattern filters). Clicking on a filter opens it
in the Active Filters area for setting parameters. Successive filters get chained sequentially; clicking on the
x next to a filter removes it from the chain. The filter combination is applied only on clicking the "Apply filters"
button.
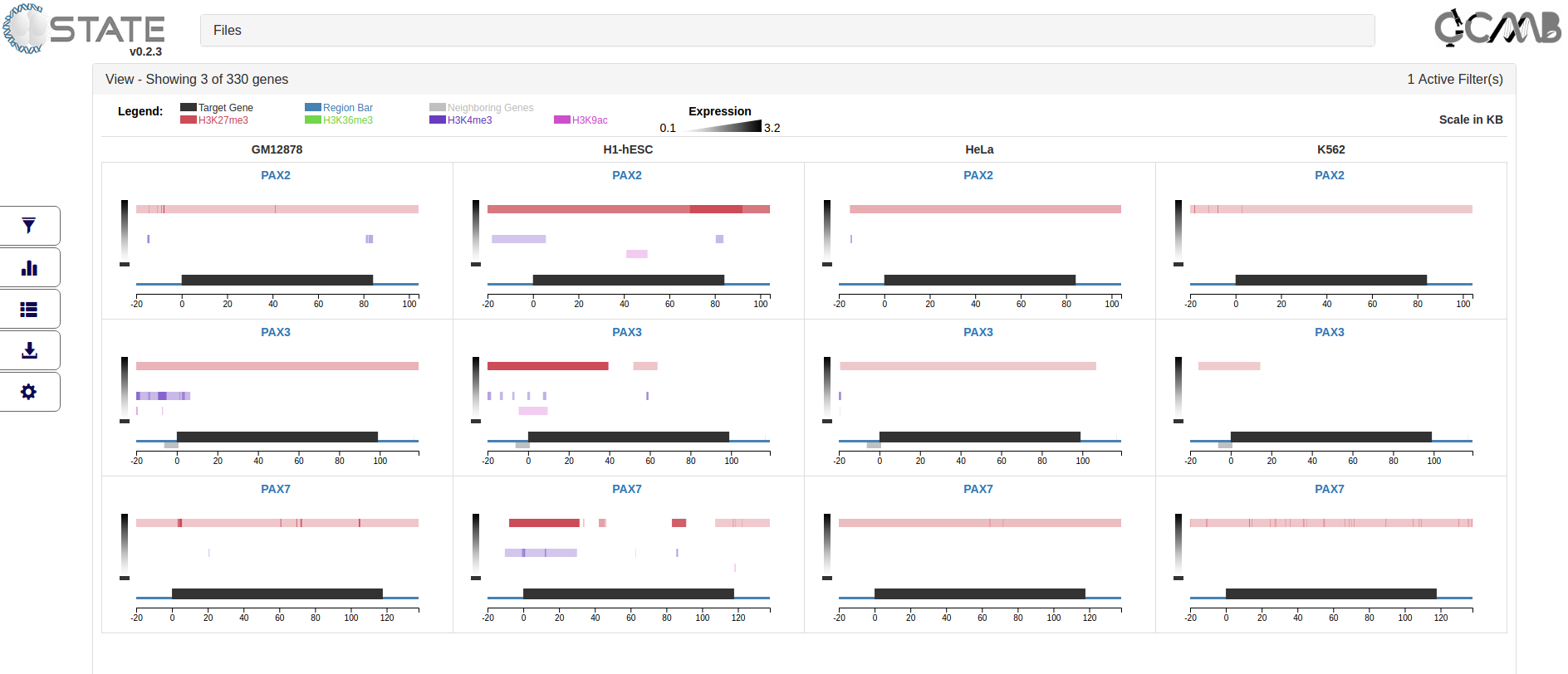
Once the filters are applied using the "Apply Filters" button, the filter panel slides back. The number of active filters
are indicated in the header of the View accordion. Filtered output can be exported and saved.
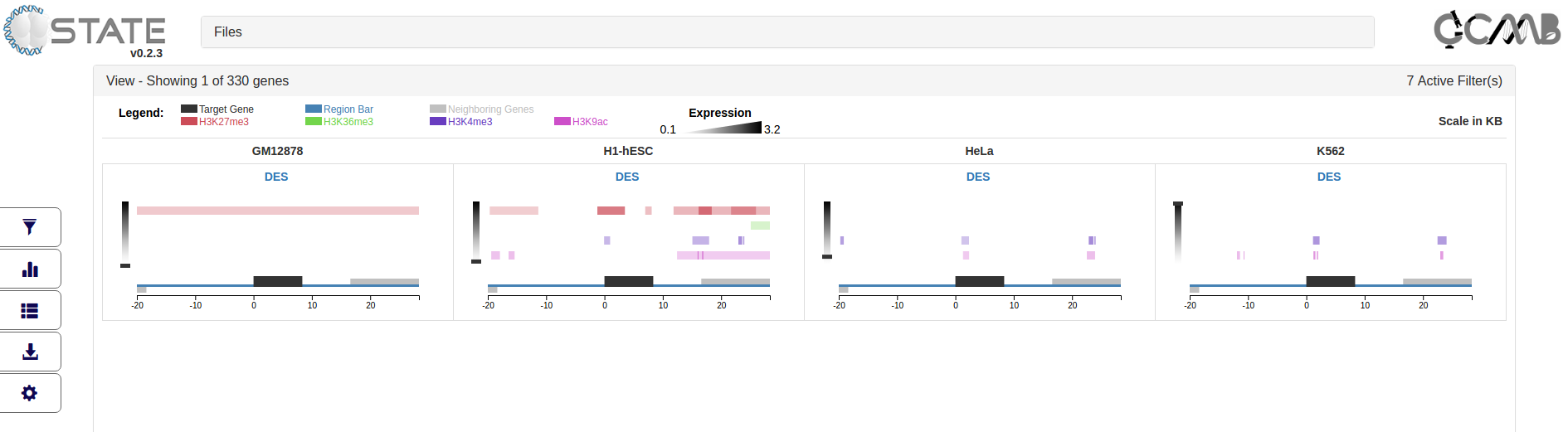
Plots and analysis panel - Identifying Data trends
In addition to gene centric plots for visualization, C-State also allows users to identify cell type specific global trends
in their dataset using the Plots and Analysis button (2nd button in the control panel). If filters are activated (from
the previous panel), the plots can be generated only on the filtered gene subsets thus enabling a platform to analyze
multiple specific versus global patterns.
Histograms
Display the frequency of features based on their size or score (X-axis) across all marks and cell types (arranged
row- and column-wise respectively by default). Use the "Switch Rows/Columns" button to toggle the arrangement of
cell types and features.
How histograms are generated...
If "Features Scores" is chosen in the dropdown menu and cell types are arranged column-wise:
- C-State retrieves the scores of all features in a given cell type
- The minimum and maximum scores of each cell type are calculated, so that the histogram bins are constant across
multiple features
- The scores are separated feature-wise, and supplied to d3.histogram with the previously calculated minimum and maximum values as data range
- Score are divided into 20 bins, and the frequency of a given feature in each bin is plotted as a bar
If rows and columns are switched, the same steps are followed, except that the minimum and maximum values are calculated
using the info of respective features across all cell types.
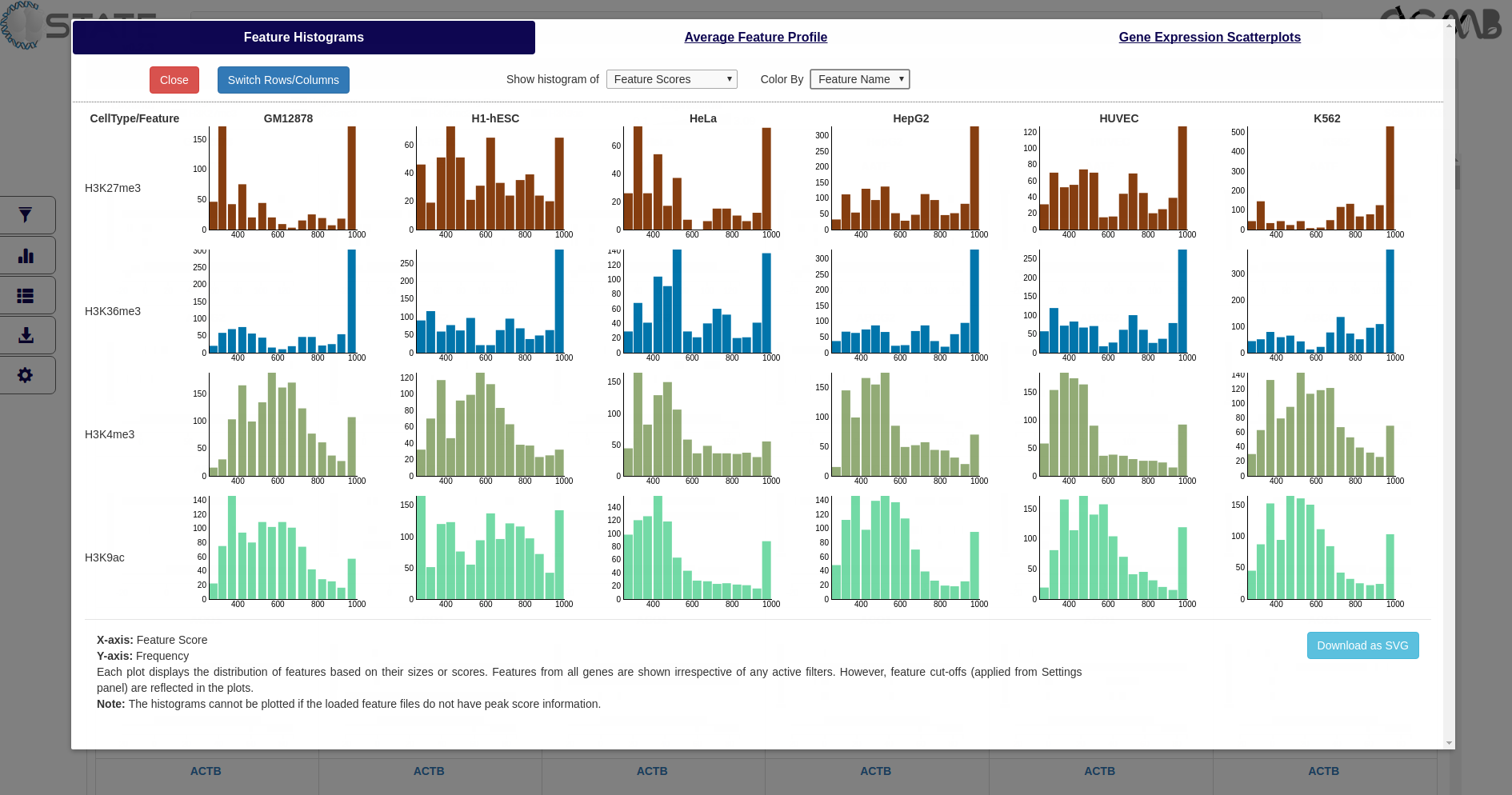 Plots and Analysis panel (Feature Histograms) showing frequency of features based on their scores (peak intensity)
and colored by feature name
Plots and Analysis panel (Feature Histograms) showing frequency of features based on their scores (peak intensity)
and colored by feature name
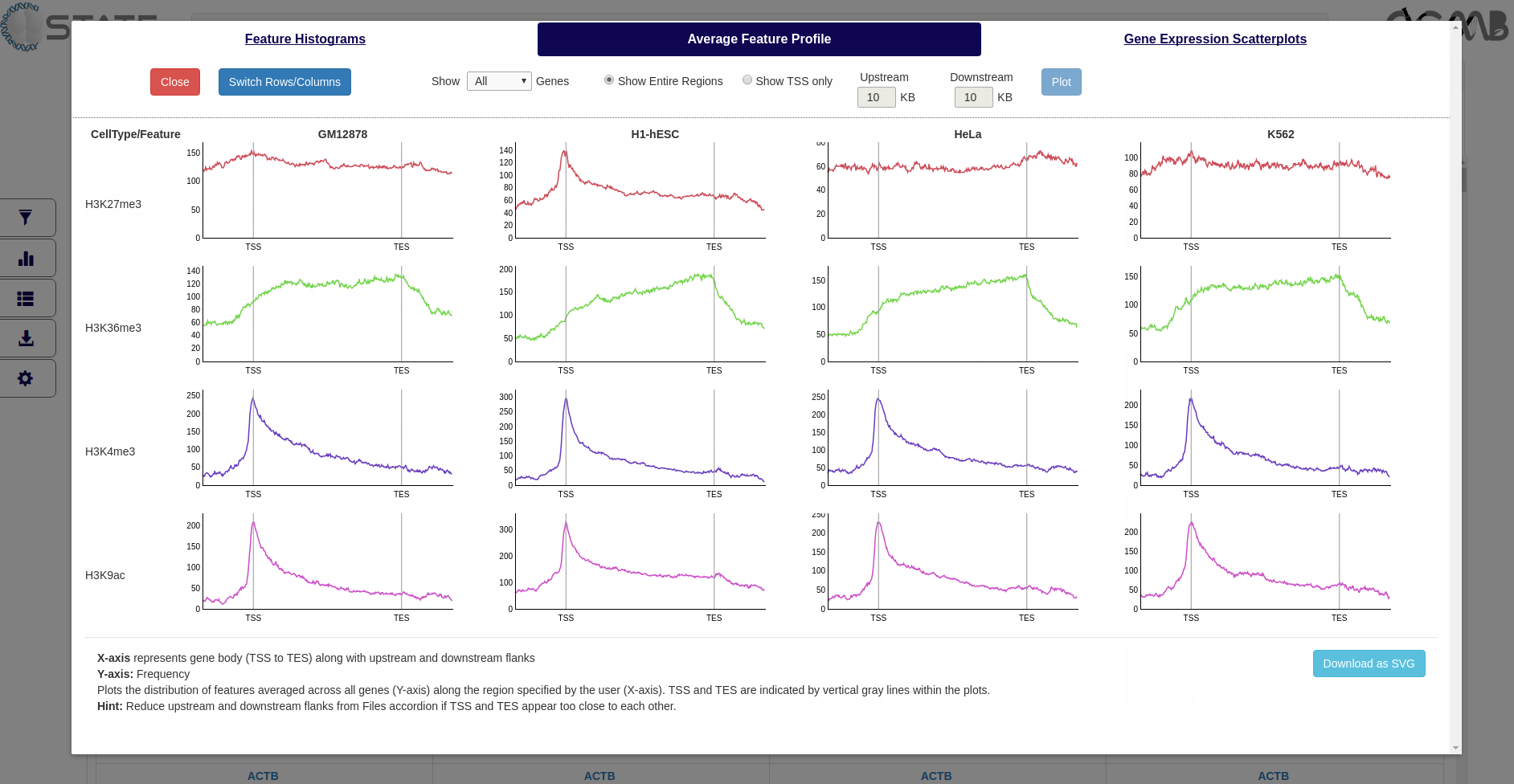
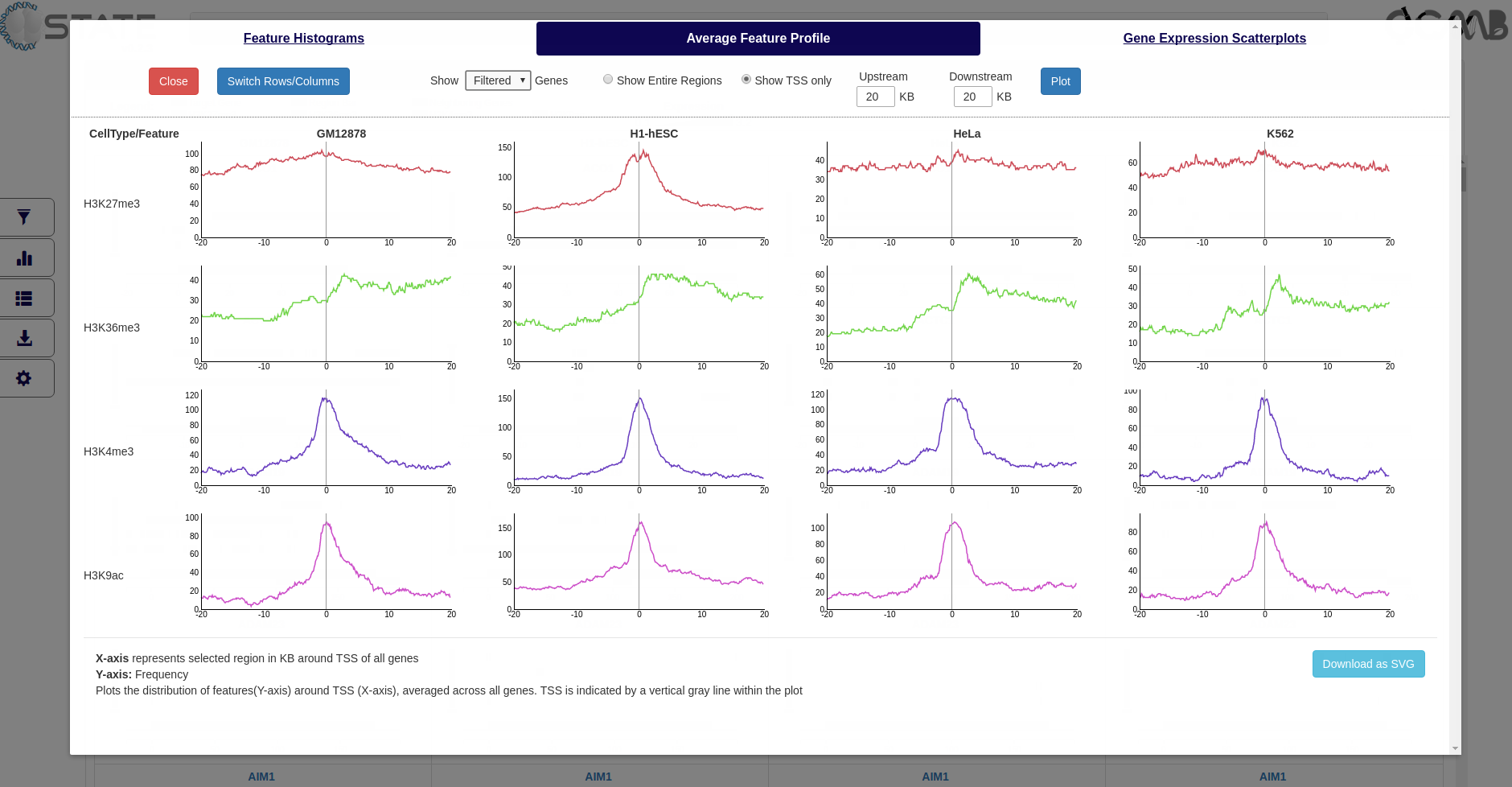
Average Feature Profile
Plot the distribution of features averaged across all genes (Y-axis) along the region specified by the user (X-axis). The
gene start and end positions and the chosen upstream and downstream flank sizes are indicated. Data can be plotted
for a filtered subset (if any filters are activated) or for the entire list of genes and the Y-axis scale can be
adjusted from the dropdowns above the plots.
How average profiles are calculated...
All genes and their respective flanks are divided into equal number of bins as follows:
- The number of gene body bins is determined by the median gene size in the list of target genes using a bin size of 100bp. For example, if the median size is 50KB, the number of gene bins is 500
- Upstream and downstream flanks are divided into 100bp bins
- Starting from the most upstream bin, C-State counts the features falling in each bin for all genes
- The frequency in each bin is plotted on Y-axis whereas the bins are represented on the X-axis
In case of TSS plots, upstream and downstream regions of TSS entered by the user are divided into 100bp bins instead of using median gene size.
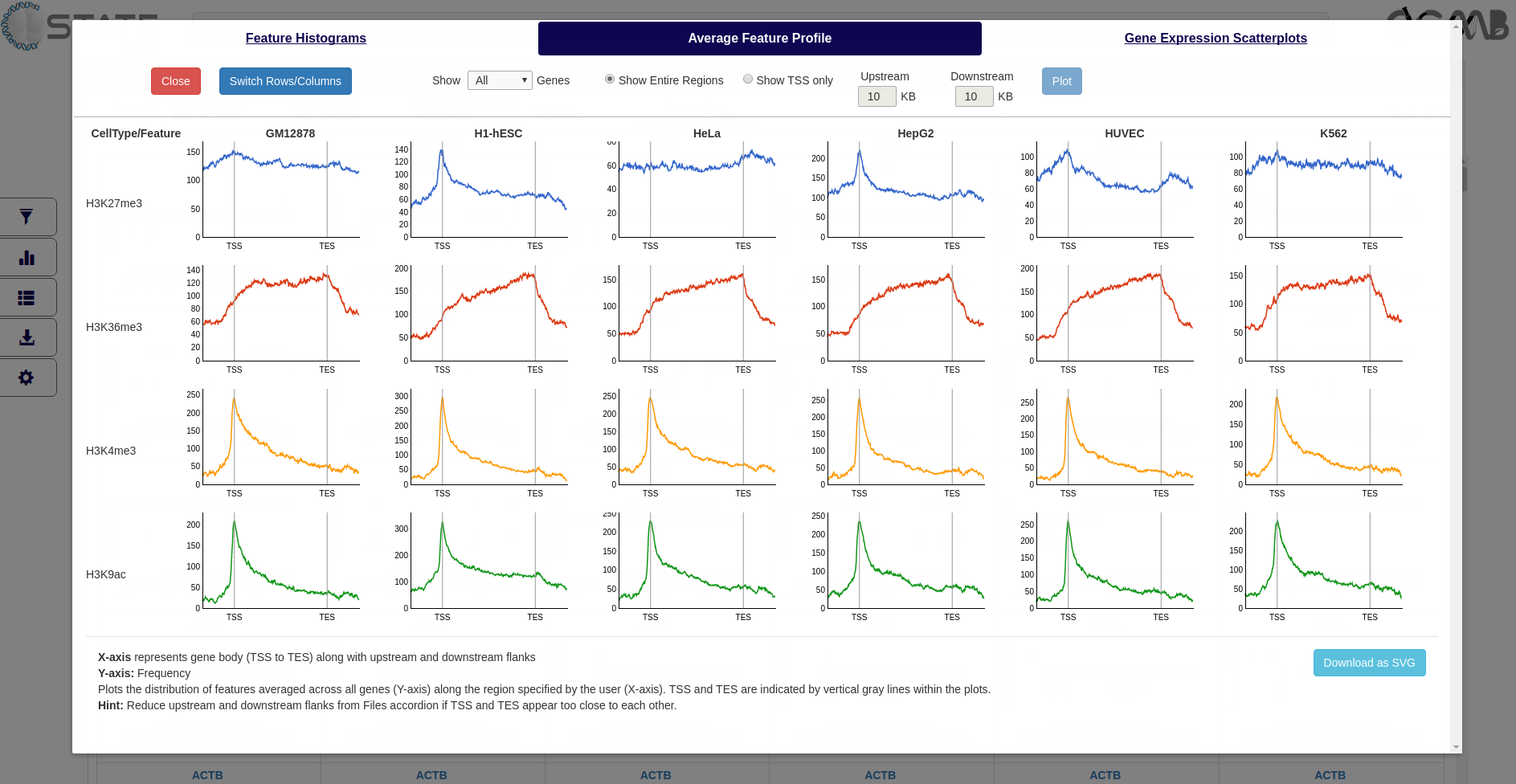 Plots and Analysis panel (Average Feature Profile) showing the average distribution of each mark with respect
to the gene bodies (TSS to TES) along with flanking regions selected in the Files accordion
Plots and Analysis panel (Average Feature Profile) showing the average distribution of each mark with respect
to the gene bodies (TSS to TES) along with flanking regions selected in the Files accordion
Gene Expression Scatterplots
Plot the relative gene expression values between pairs of cell types. Expression level of a gene in the first cell type (column name) is plotted on X-axis, and its expression in the second cell type (row name) is plotted on Y-axis.
How gene expression range is determined...
- C-State calculates the cumulative minimum, maximum, and the 5th, 95th, and 99th percentile of expression values from the expression data files of all the cell types
- By default, the axis minimum and maximum are determined by the minimum and 99th percentile of the loaded datasets
- These values can be changed using the axis input boxes on the top of the plots
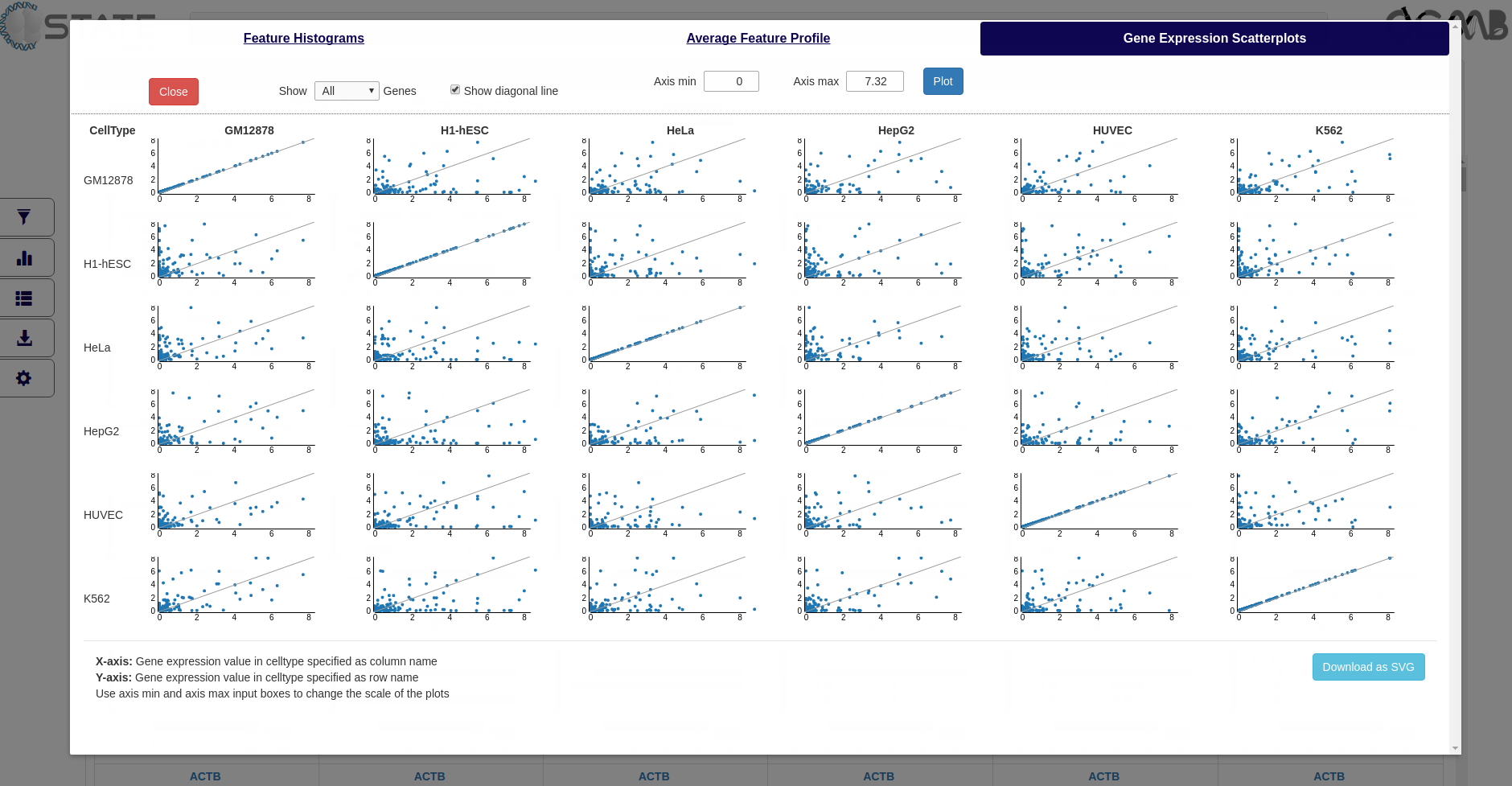 Plots and Analysis panel (Gene Expression Scatterplots) showing comparative distribution of gene expression profiles for all target genes
Plots and Analysis panel (Gene Expression Scatterplots) showing comparative distribution of gene expression profiles for all target genes
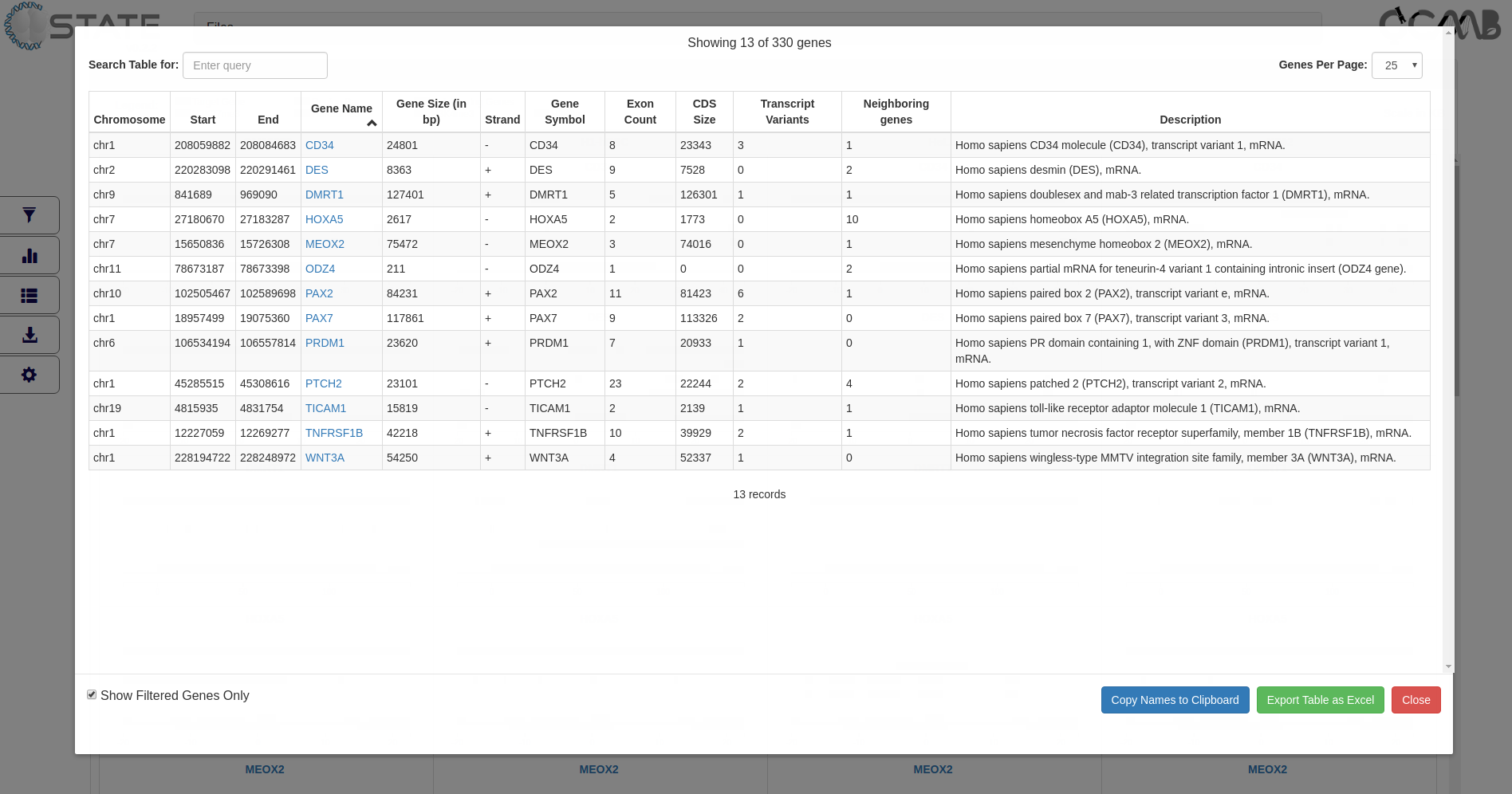
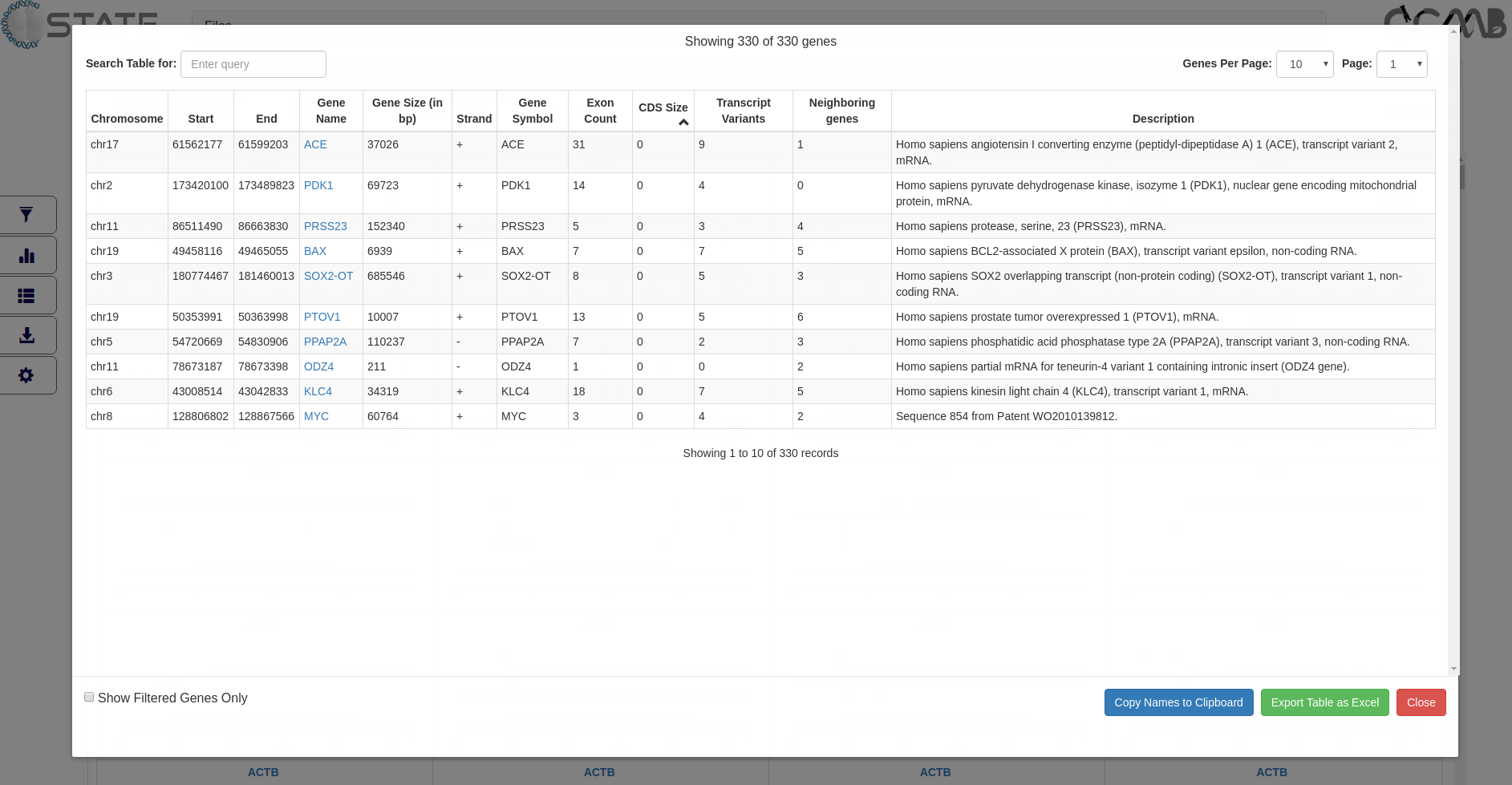
Tables panel - View tabular data
Tabular information (3rd button in the control panel) of the gene set is provided to display gene information of all or filtered
subsets of genes (if filters are activated). The table is interactive and can be sorted on any of the columns. Clicking
on the gene name opens the gene modal view for that gene. This feature is useful for sorting information based on
user-defined criteria and subsequently viewing the gene panels in the desired order.
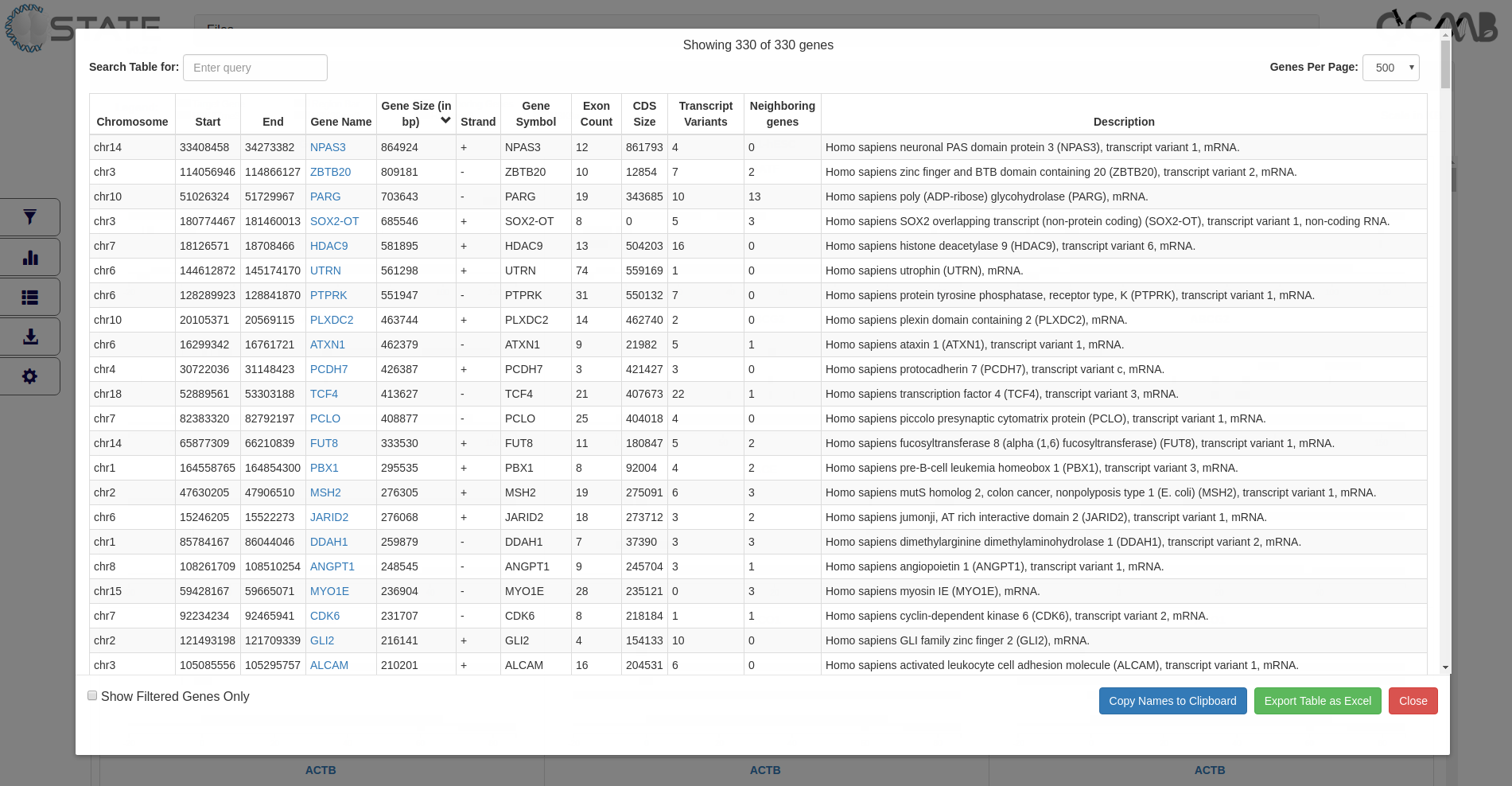 Tables panel showing tabulated gene information of all target genes
Tables panel showing tabulated gene information of all target genes
The entries can be searched based on terms in any of the columns and arranged page wise using the controls at the top
of the table. Clicking the “Copy to clipboard” option copies all the gene names displayed in the table for handy
pasting into any other application or relevant rows may directly be copied from the table. Selected pages or the
entire table can be exported and saved.
Downloads panel - Download Results
The Downloads button (4th button in the control panel) provides 3 options to save and export the various outputs generated
in C-State.
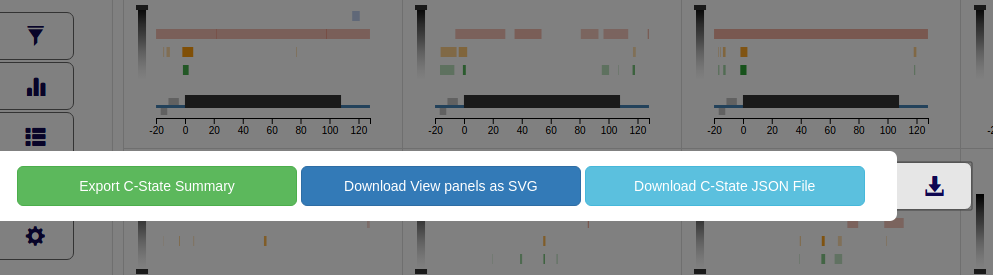 Downloads panel showing options to download multiple C-State outputs
Downloads panel showing options to download multiple C-State outputs
Export C-State Summary - Export a txt file containing summary of the gene names, feature track settings, and active filters (if any).
Download View panels as SVG - Download all the gene panels in the View accordion as a single SVG file.
Download C-State JSON File - Download the entire session info and plot data as a JSON file. This file can be uploaded as "Previous C-State Data" from the Files accordion.
Settings panel - Change global settings
Settings changed from this panel apply across C-State, for visualization, filtering, plotting and analysis and also
get saved in any exported / downloaded files.
Feature Tracks
Feature track settings can be changed to define the quality of the peaks to be displayed (based on feature size and/or score).
The user can provide cut-offs to ensure that only peaks meeting the selected criteria are displayed. The color
of each of the feature tracks can be selected from 5 color schemes.
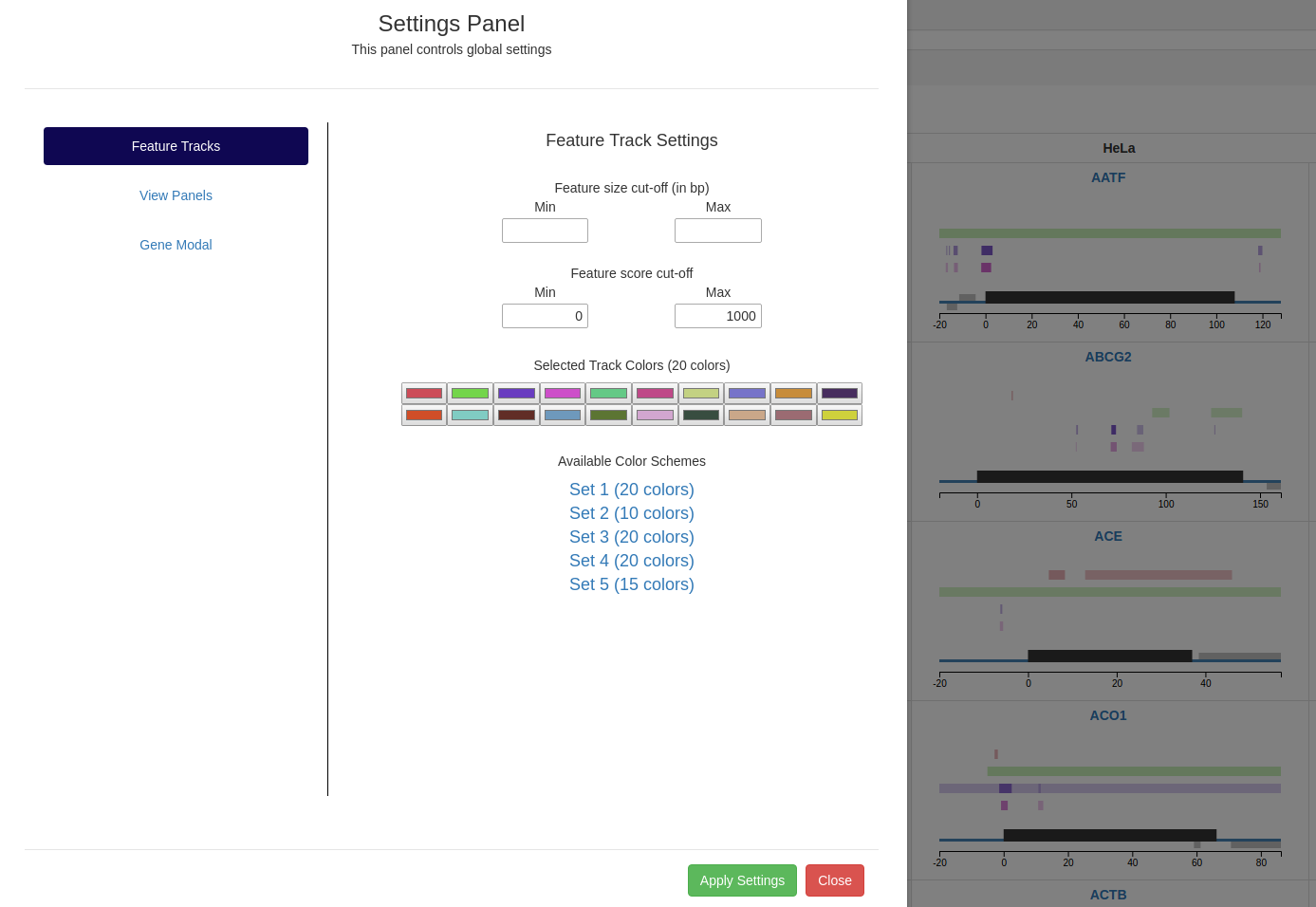 Settings panel showing options to change feature track attributes
Settings panel showing options to change feature track attributes
View Panels
Feature bar attributes (height, color) can be defined by the user. By default, exons are not displayed in the main view gene
panels but they can be set for display here, along with neighboring genes. The range (default is 5th and 95th percentile) and color palette (default grayscale) used to represent the gene expression level can be customized.
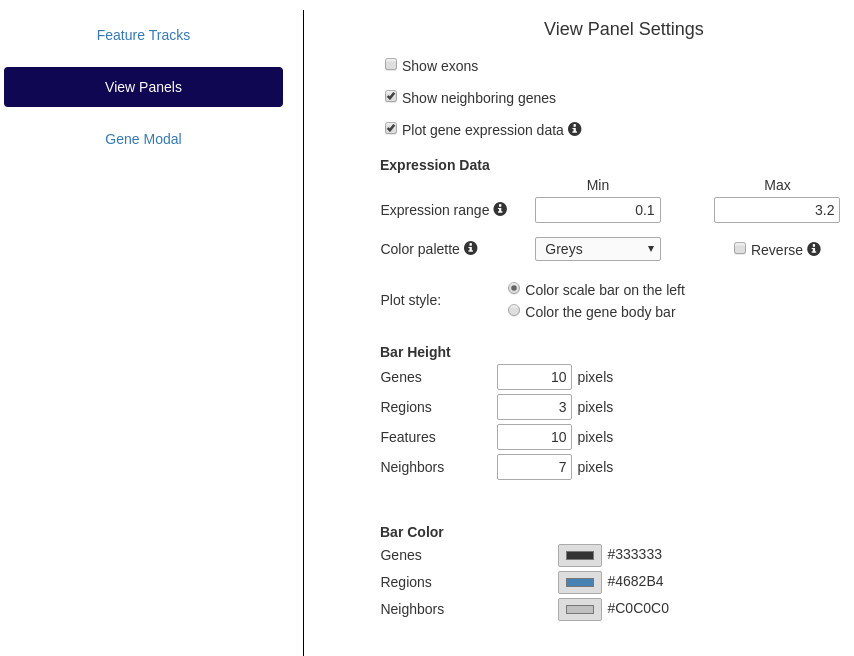 Settings panel showing options to change view panel attributes
Settings panel showing options to change view panel attributes
Gene Modal
Similar settings as in the main view are available here. These can be changed independently or linked to the settings in
the main view.
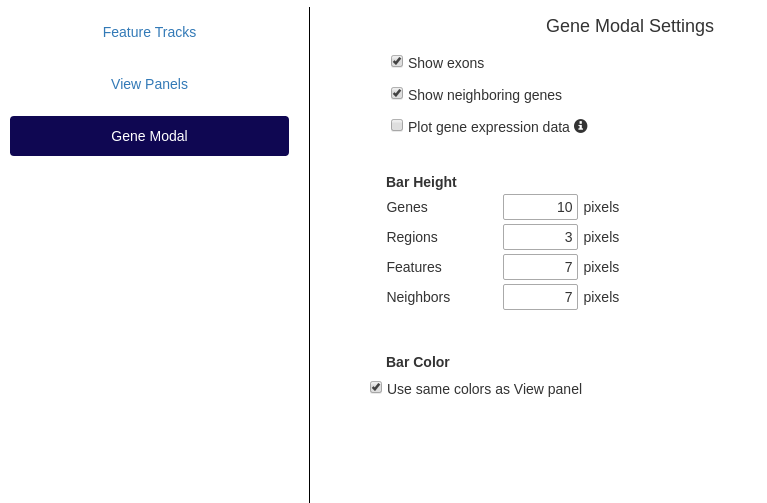 Settings panel showing options to change gene modal attributes
Settings panel showing options to change gene modal attributes
Default Colors for C-State
| Component |
Color |
Hex Value |
RGB Value |
| Gene Bar |
Black |
333333 |
51,51,51 (Hue = 0) |
| Region Bar |
Steel Blue |
4682B4 |
70,130,180 |
| Neighbors |
Silver |
C0C0C0 |
192,192,192 |