Some of the ongoing projects in the lab are as follows.........
Structural studies of dsRBD containing proteins in the RNAi pathway
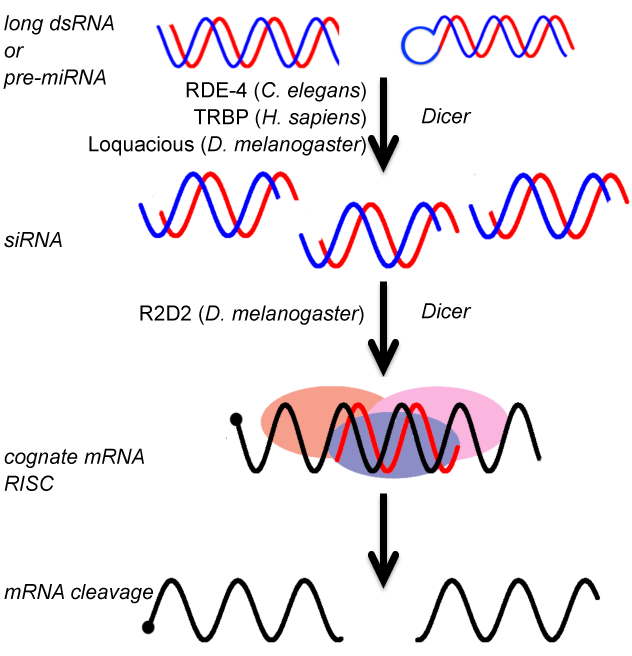
The key steps in any RNA interference pathway are cleavage of dsRNA or pre-miRNA into siRNA/miRNA, loading of the anti-sense strand onto the RISC complex and cognate mRNA silencing mediated by the RISC complex. Two key enzymes, Dicer and Argonaute, are assisted by a variety of multiple dsRNA binding domains (dsRBDs) containing proteins (dsRBPs) to effect the pathway. Although the RNAi pathway appears highly conserved in higher eukaryotes subtle differences in assisting proteins often alters the fate of this important post-transcriptional gene regulation pathway.
In organisms like C. elegans and H. sapiens, only one Dicer and one or two dsRBDs are found to dictate both siRNA and miRNA biogenesis. However, organisms like D. melanogaster have two separate sets of Dicer:dsRBPs to execute both siRNA and miRNA pathways. In the other extreme, A. thaliana encodes four Dicers and four dsRBPs to carry out siRNA and miRNA driven gene silencing.
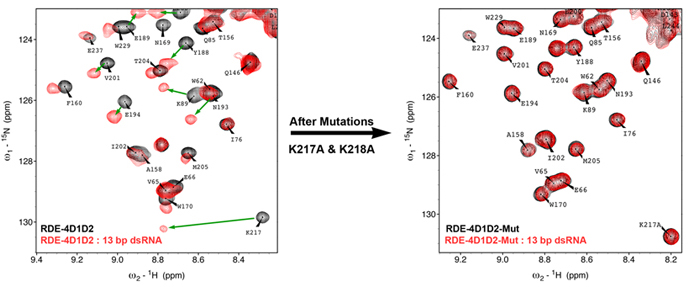 Interestingly, bioinformatics analyses of sequences of these Dicer and dsRBPs show a variety of differences, for instance, the linker between RDE-4 dsRBDs in C. elegans is composed of ~ 70 amino acids, whereas, in the majority of its counterparts the linker length spans only 5-10 amino acids. Several prominent changes in terms of C-terminal regions, dimerization etc. can be further noticed. Various Dicer domains are also seen to possess major modifications.
Interestingly, bioinformatics analyses of sequences of these Dicer and dsRBPs show a variety of differences, for instance, the linker between RDE-4 dsRBDs in C. elegans is composed of ~ 70 amino acids, whereas, in the majority of its counterparts the linker length spans only 5-10 amino acids. Several prominent changes in terms of C-terminal regions, dimerization etc. can be further noticed. Various Dicer domains are also seen to possess major modifications.
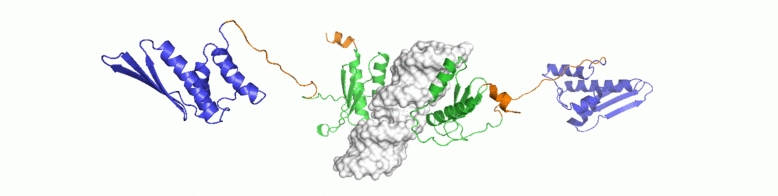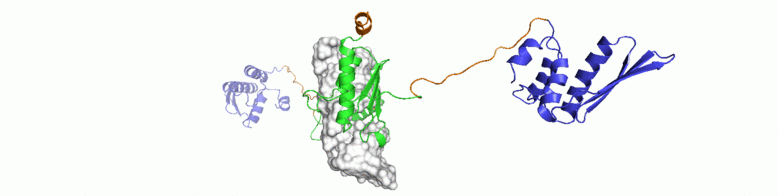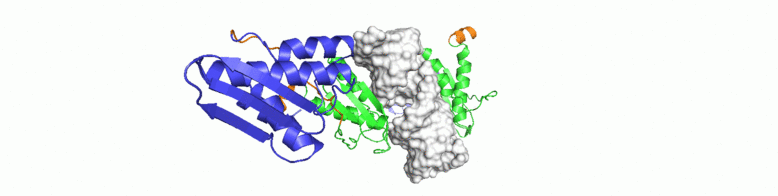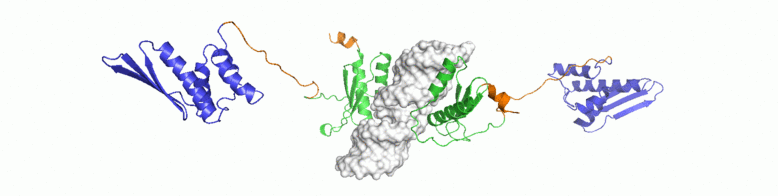
For a conserved and ancient mechanism like RNAi, why would organisms need to alter key enzymes and proteins? At what stage, have the key components diverged? We explore answers to some of these questions through NMR driven solution structures of dsRBPs from C. elegans and D. melanogaster as well as study their interactions with dsRNA and minimal interaction domains of Dicer.
Crc as a global post-transcriptional gene regulator in Pseudomonas
Carbon Catabolite Repression (CCR) allows bacteria to selectively assimilate a preferred compound among a mixture of several potential carbon sources, thus boosting their ability to survive. Crc (Catabolite Regulation Control), an RNA-binding-protein, is a global regulator in CCR in Pseudomonas. Crc (~31 kDa) triggers a repression process that inhibits the expression of genes involved in transport and catabolism of non-preferred substrates, thus indirectly favoring assimilation of preferred ones. A dissection of the molecular mechanisms underlying CCR function is vital to comprehend how biodegradation is performed by Pseudomonads in their natural environmental niche. We are addressing this issue by determining the solution structure of Crc and subsequently identifying RNA binding sites using NMR-based titrations. The work will also explore possibilities of conformational exchange processes involved in RNA recognition by Crc.
How does RBP42RRM recognize coding regions of mRNA to effect gene silencing?
RNA binding proteins (RBPs) have been known to play a significant role in the mRNA metabolism of diverse organisms such as C. elegans, D. melanogaster and T. brucei etc. RBPs have a modular structure comprised of multiple repeats that are built from a number of RNA binding domains, which bind to untranslated regions of mRNA. RBPs assist the processing and assembly of non-coding RNAs into ribonucleoprotein complexes, which mediate important cellular functions such as splicing and translation.
In T. brucei, a novel RNA binding protein named RNA binding protein 42 (RBP42) of ~ 38 kDa has been recently identified (Das et al. RNA 2012) and has been hypothesized to play a major role in adaptation to different environment by regulating mRNA metabolism. RBP42 targets the coding region of mRNA that encodes proteins involved in cellular energy
metabolism and shows similarity to the mammalian RNA binding protein, G3BP. The N terminal region of RBP42 has homology to the Nuclear transport factor (NTF2- like protein) and C terminus with the RNA recognition motif (RRM). Structural studies of full length RBP42 and its domains as well as RNA binding studies are in progress.
How do multidomain protein organize to perform sychronous functions?
Multidomain proteins often show interesting yet complex dynamic behaviour. Inter-domain motions are crucial to substrate recognition and prevail over the static structure or fold. Dynamic fluctuation in conformations occuring at different magnitudes and timescales are often functionally correlated. In RNAi, formation of a stable heteromeric complex between Dicer:dsRBP and dsRNA induces intense conformational dynamics on both, the protein and enzyme. Indications on the ms-μs dynamics have been already observed for some of these proteins. Solution NMR spectroscopy is a uniquely suited technique to obtain information on order parameters and exchange processes at the atomic level details.
We are studying 15N relaxation rates and CPMG to investigate conformational exchange processes in dsRNA recognition of RDE-4. Moreover, using paramagnetic relaxation as well as internal alignment of individual domains, we are in the process of obtaining a
comprehensive model of motional averaging in the dsRBPs and their complexes with dsRNA.
 Interestingly, bioinformatics analyses of sequences of these Dicer and dsRBPs show a variety of differences, for instance, the linker between RDE-4 dsRBDs in C. elegans is composed of ~ 70 amino acids, whereas, in the majority of its counterparts the linker length spans only 5-10 amino acids. Several prominent changes in terms of C-terminal regions, dimerization etc. can be further noticed. Various Dicer domains are also seen to possess major modifications.
Interestingly, bioinformatics analyses of sequences of these Dicer and dsRBPs show a variety of differences, for instance, the linker between RDE-4 dsRBDs in C. elegans is composed of ~ 70 amino acids, whereas, in the majority of its counterparts the linker length spans only 5-10 amino acids. Several prominent changes in terms of C-terminal regions, dimerization etc. can be further noticed. Various Dicer domains are also seen to possess major modifications.

