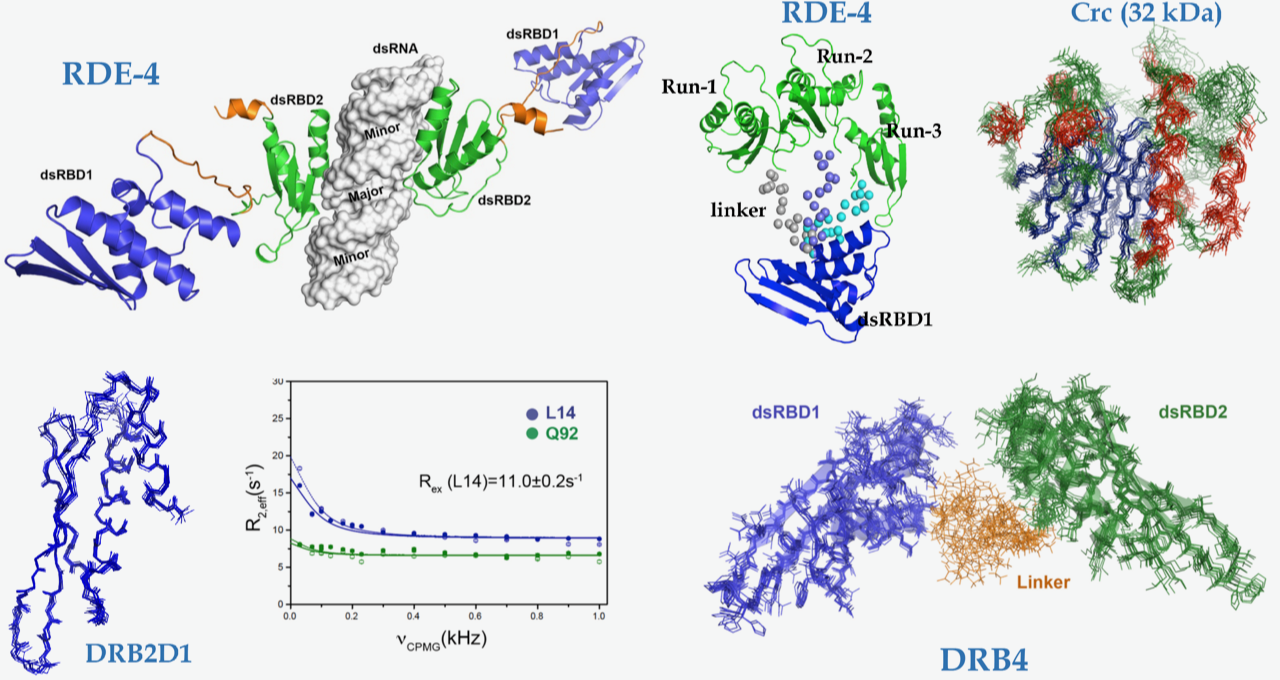
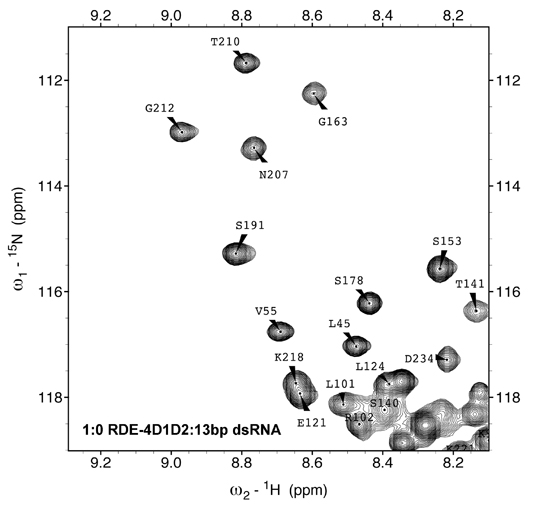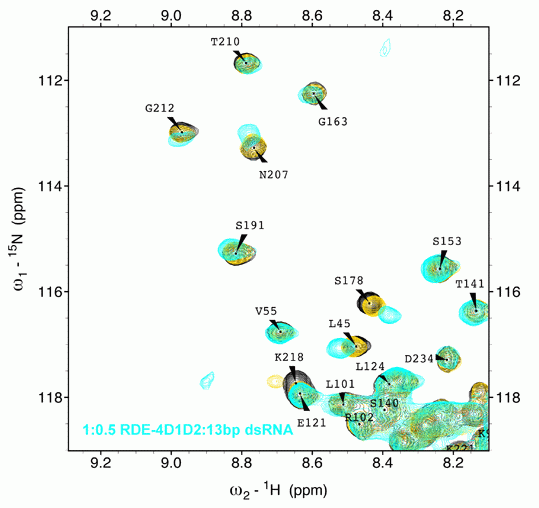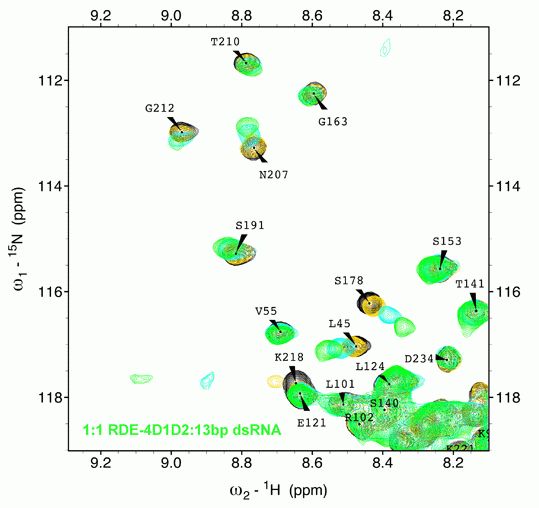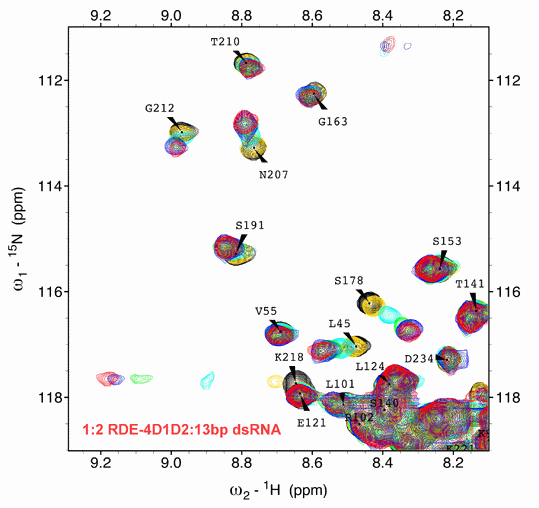
We explore the role of regulatory proteins that bind to a variety of RNA molecules and achieve gene regulation. In this direction, the group investigates the structural and mechanistic details of various components of the RNAi machinery amenable to solution NMR techniques. We are involved in understanding the mechanism of Dicers:dsRBPs complex in various organisms ranging from C. elegans to A. thaliana. Results obtained from our lab and others elaborate on the structural divergence in seemingly conserved and highly homologous proteins which implies a fine balance in which subtle changes can make or break the RNA mediated gene silencing. Our results further exemplify that the process of RNAi initiation is unique for each organism and is heavily dependent on a step-wise assembly of the Dicer, its partner proteins, and the trigger small RNA.
Additionally, we probe structure-function relationships in regulatory RNAs such as riboswitches and RNA binding proteins in prokaryotes. The lab utilizes solution NMR spectroscopy as a major tool together with complementary techniques in molecular biology, biochemistry, and biophysics.
webmaster: mvdesh@ccmb.res.in
Last updated: May 2025